Recommendations for accurate genotyping of SARS-CoV-2 using amplicon-based sequencing of clinical samples - ScienceDirect

Difference between sequencing Coverage and depth. Depth vs Coverage. Why they are important? - YouTube
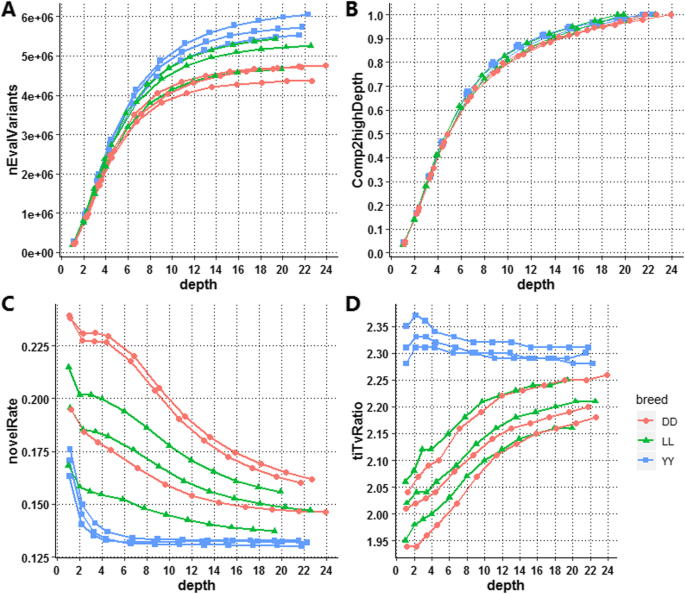
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text

Mean mapped depth and coverage of diagnostic genomic regions according... | Download Scientific Diagram

Sample depth of coverage. Histogram of the mean sequencing read depth... | Download Scientific Diagram

Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers
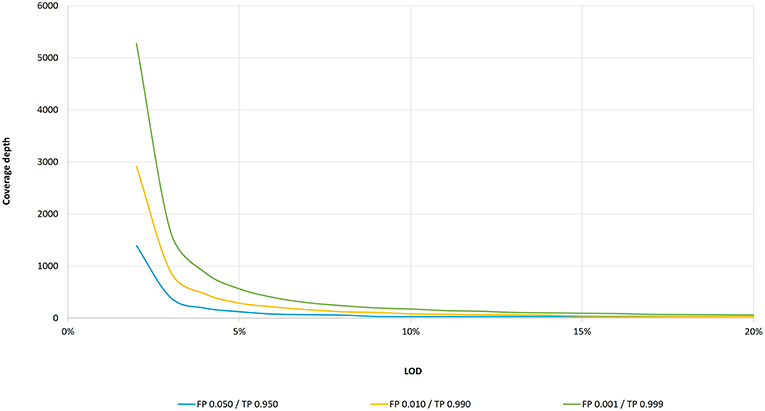
Frontiers | Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics


![PDF] Modeling genome coverage in single-cell sequencing | Semantic Scholar PDF] Modeling genome coverage in single-cell sequencing | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a8aee5ceca56b097725fb17dd908e66dd4e851de/2-Figure1-1.png)