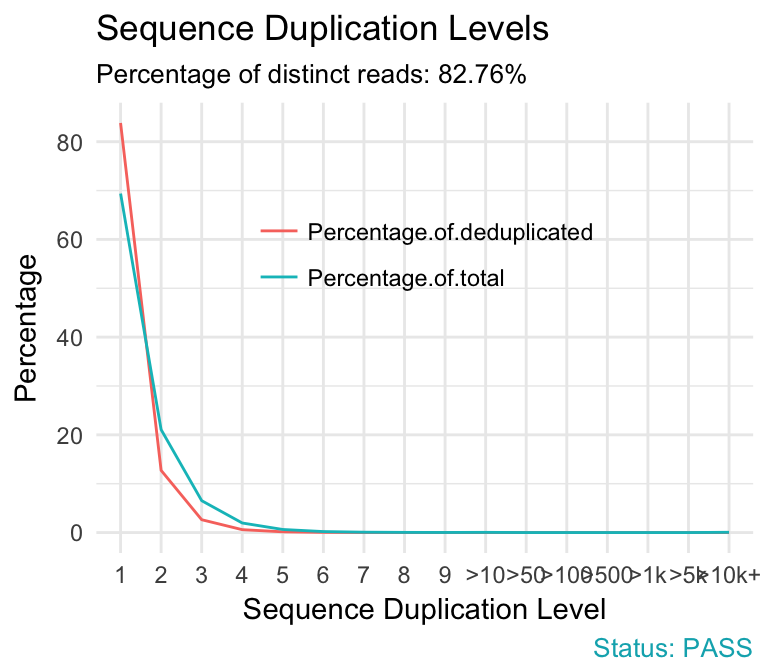
fastqcr: An R Package Facilitating Quality Controls of Sequencing Data for Large Numbers of Samples | R-bloggers
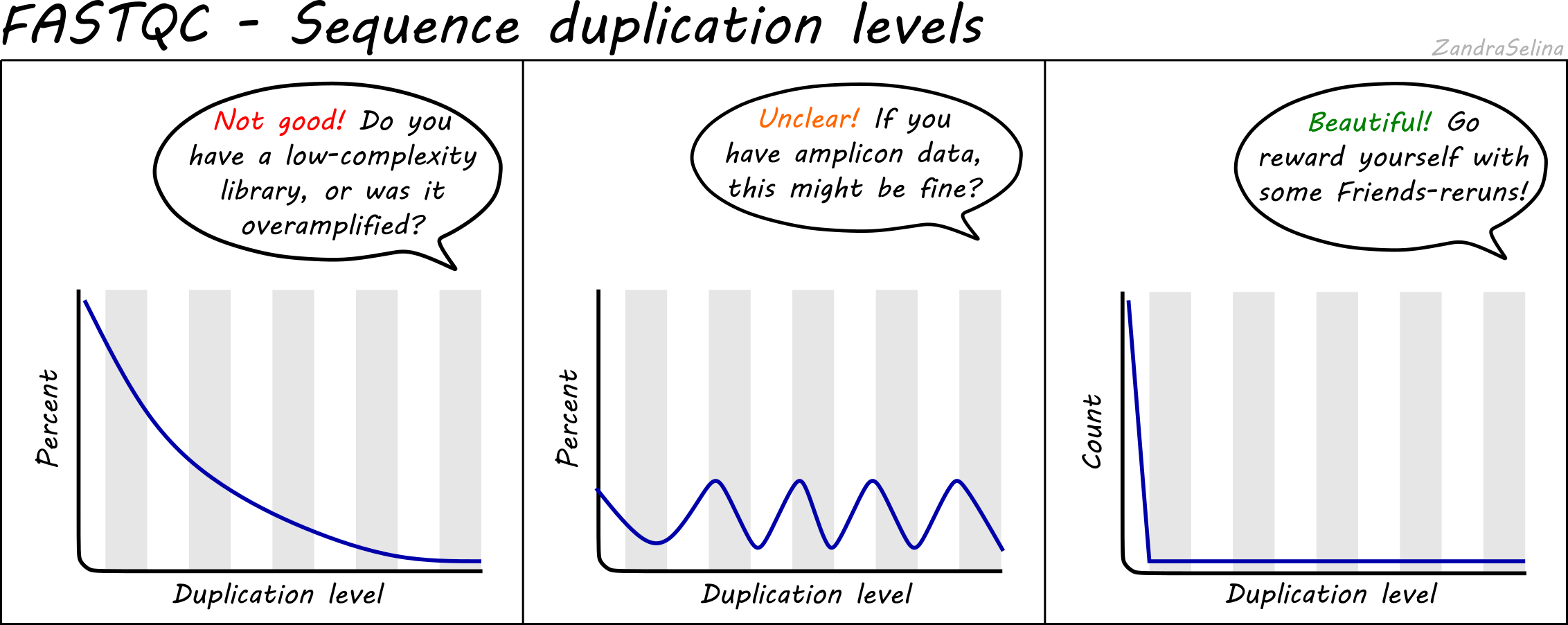
FastQC for RNA-Seq-Fails for per base seq content, gc content, duplication levels, adapter content and kmer content? | ResearchGate

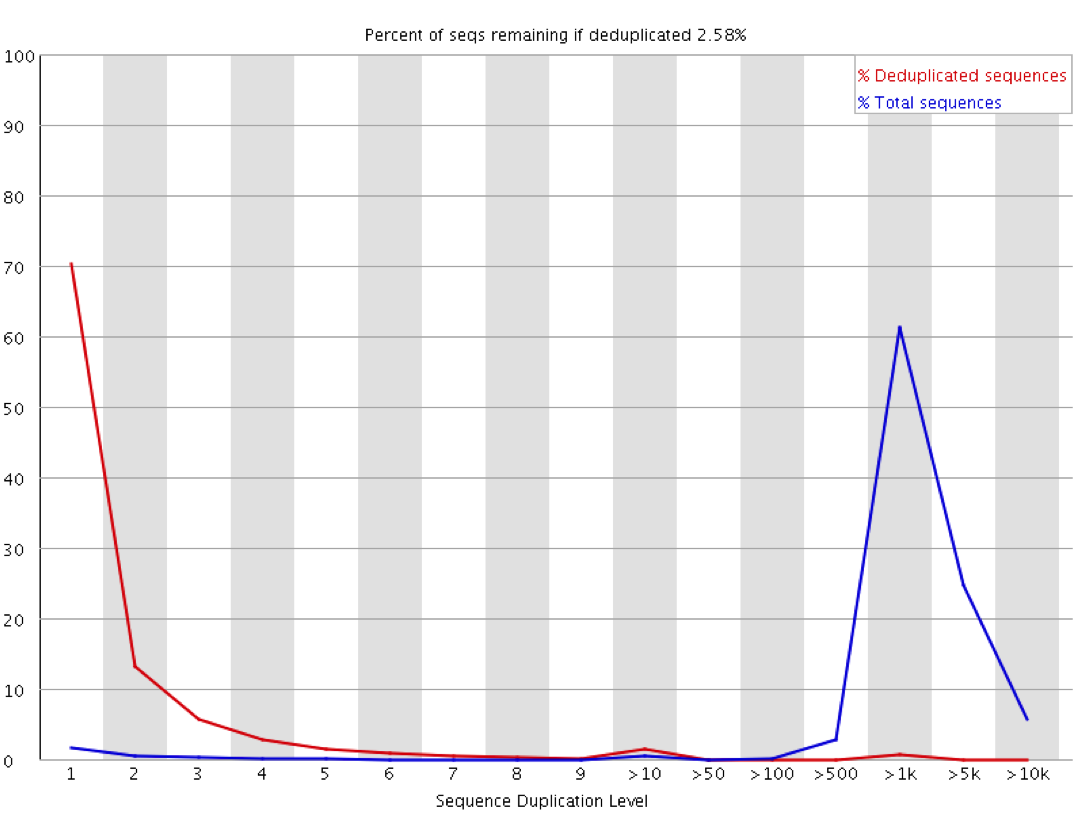
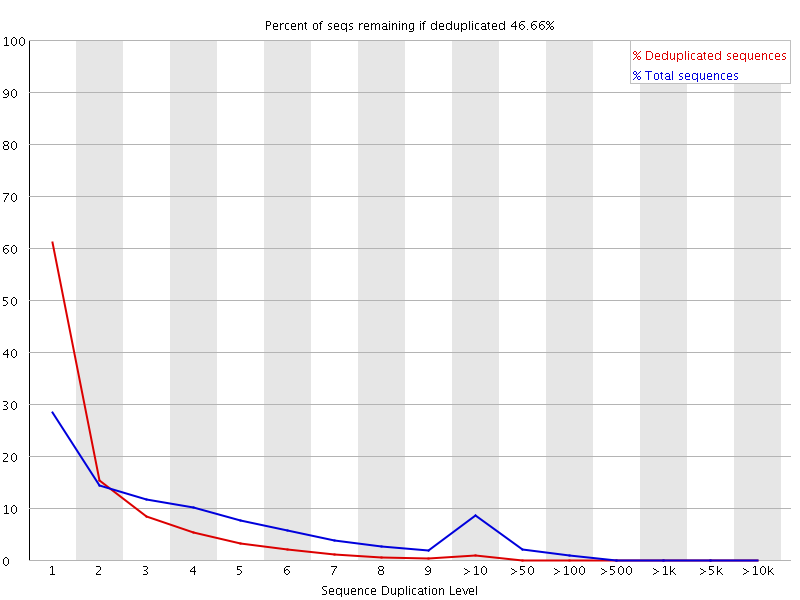
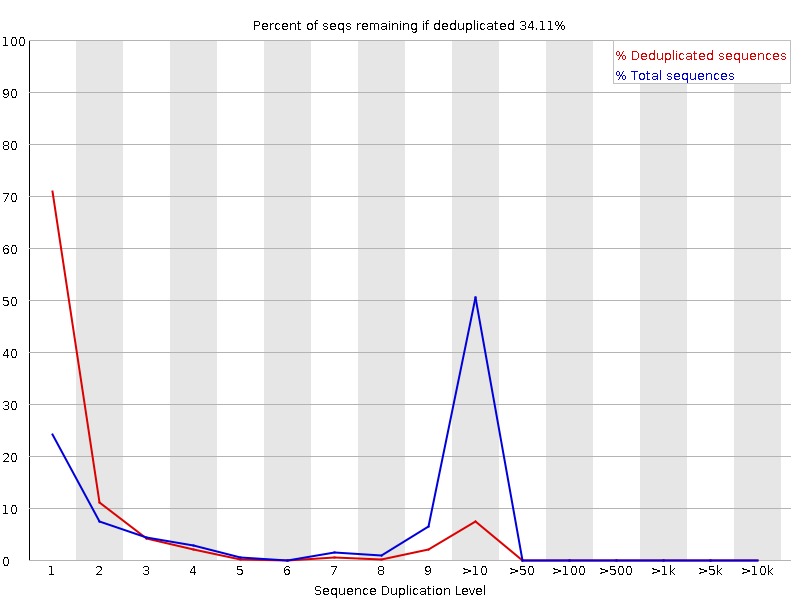
Duplication rate differs up to 30% from that of Fastqc for single end reads · Issue #290 · OpenGene/fastp · GitHub
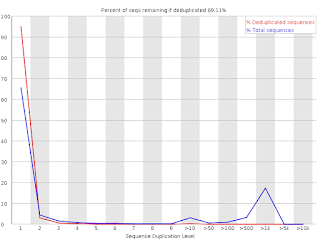
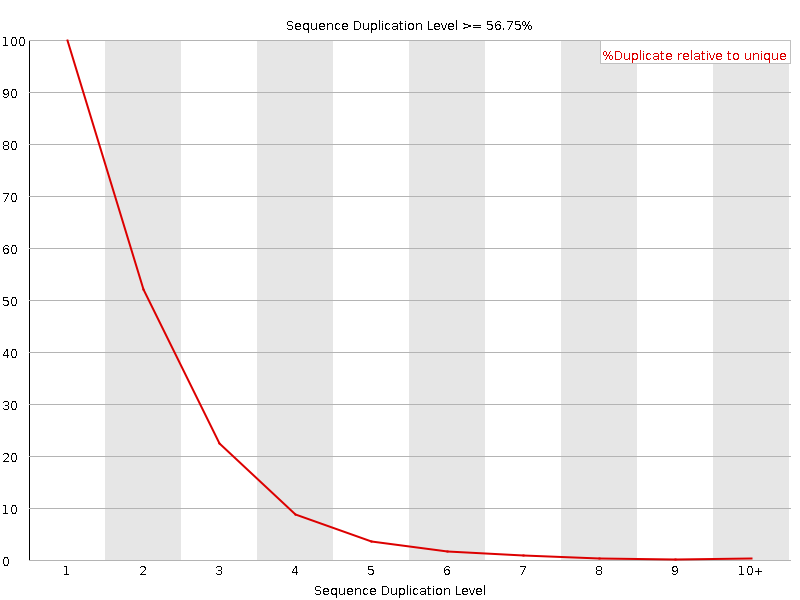
Increased read duplication on patterned flowcells- understanding the impact of Exclusion Amplification - Enseqlopedia

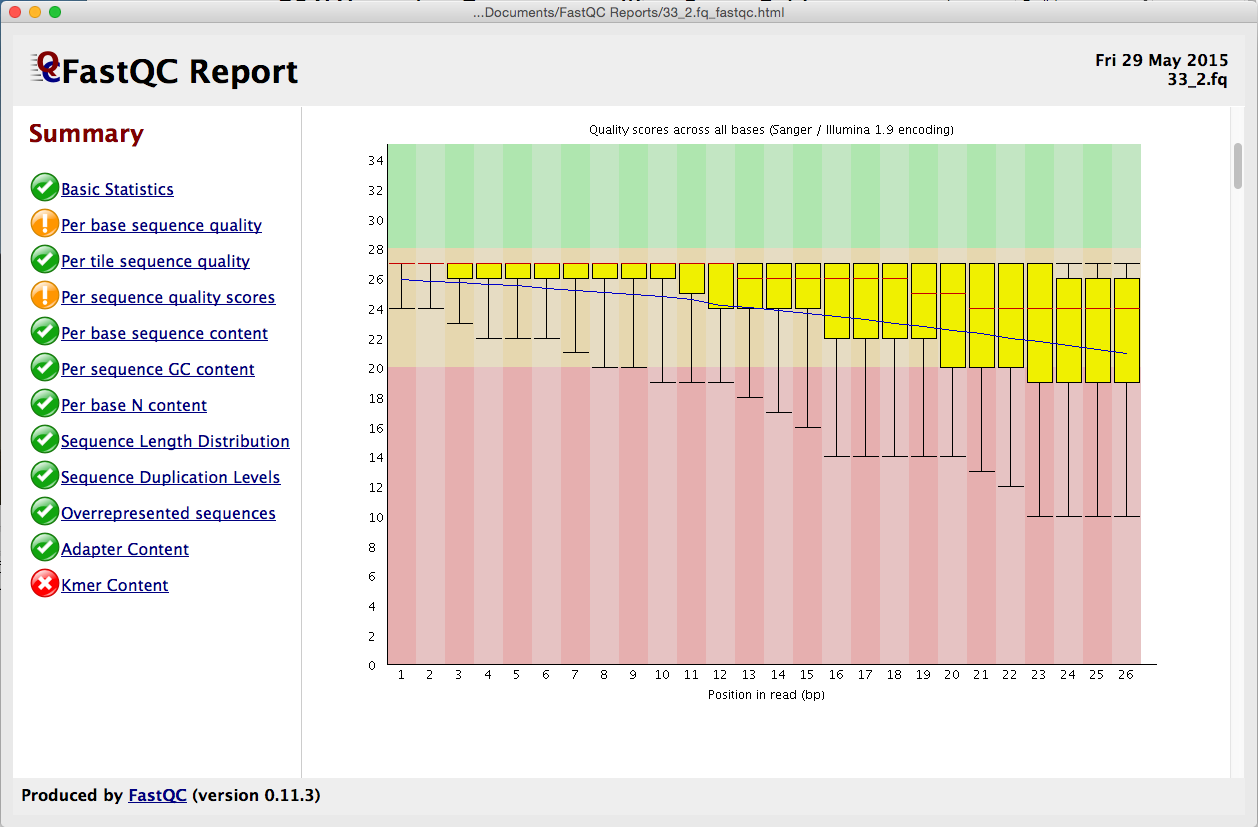
FASTQ Quality Assurance Tools - Bioinformatics Team (BioITeam) at the University of Texas - UT Austin Wikis
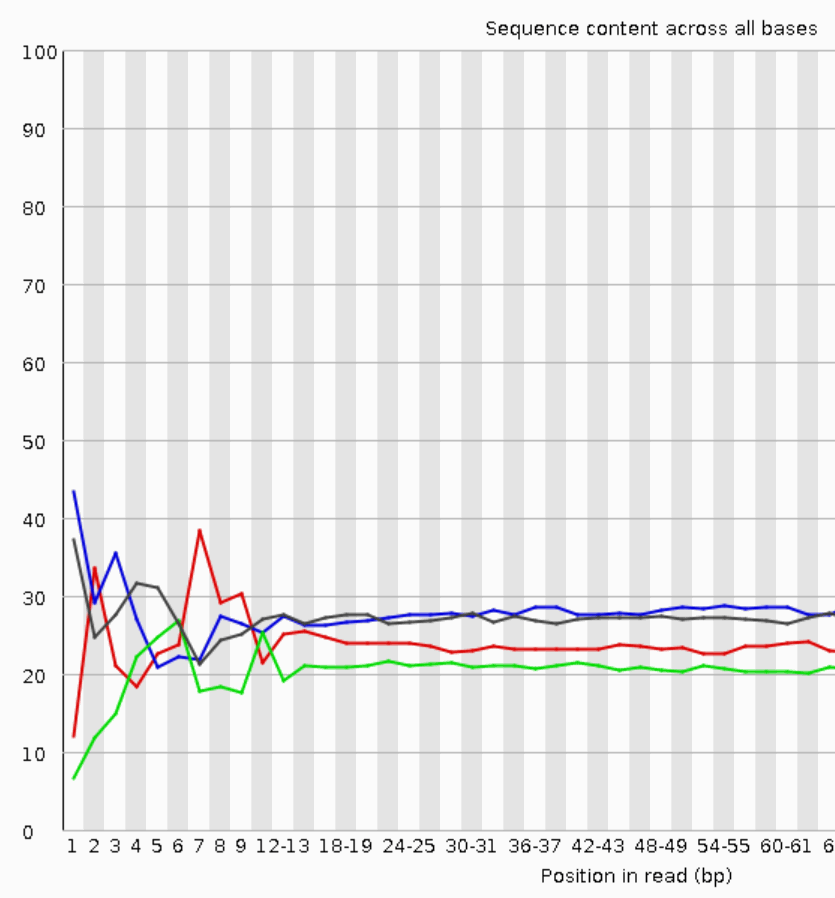
Why does FASTQC show unexpectedly high sequence duplication levels (PCR-duplicates)? | DNA Technologies Core