
Discovery of N-(4-(6-Acetamidopyrimidin-4-yloxy)phenyl)-2-(2-(trifluoromethyl)phenyl)acetamide (CHMFL-FLT3-335) as a Potent FMS-like Tyrosine Kinase 3 Internal Tandem Duplication (FLT3-ITD) Mutant Selective Inhibitor for Acute Myeloid Leukemia ...

Follistatin is a novel therapeutic target and biomarker in FLT3/ITD acute myeloid leukemia | EMBO Molecular Medicine
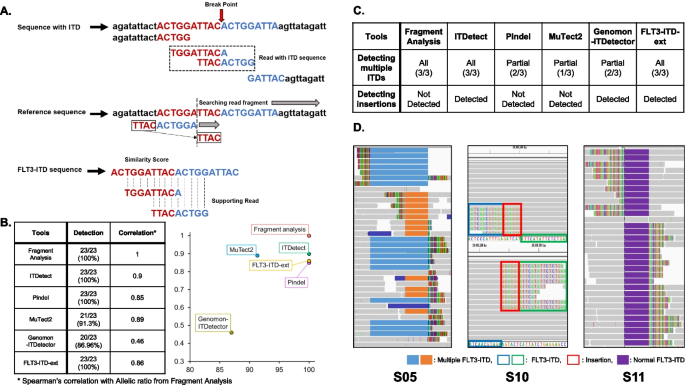
ITDetect: a method to detect internal tandem duplication of FMS-like tyrosine kinase (FLT3) from next-generation sequencing data with high sensitivity and clinical application | BMC Bioinformatics | Full Text

Prediction of HLA-binding peptides encoded by the FLT3-ITD sequence... | Download Scientific Diagram
Targeting Oncoprotein Stability Overcomes Drug Resistance Caused by FLT3 Kinase Domain Mutations | PLOS ONE

Sequence analysis of exons 14 and 15 of FLT3 gene. Inserted nucleotides... | Download Scientific Diagram
FLT3 internal tandem duplication mutations in adult acute myeloid leukaemia define a highâ•'risk group

Amino acid sequences duplicated by FLT3/ITD. At the top, amino acid... | Download Scientific Diagram
Incidence of internal tandem duplications and D835 mutations of FLT3 gene in chronic myeloid leukemia patients from Southern Ind
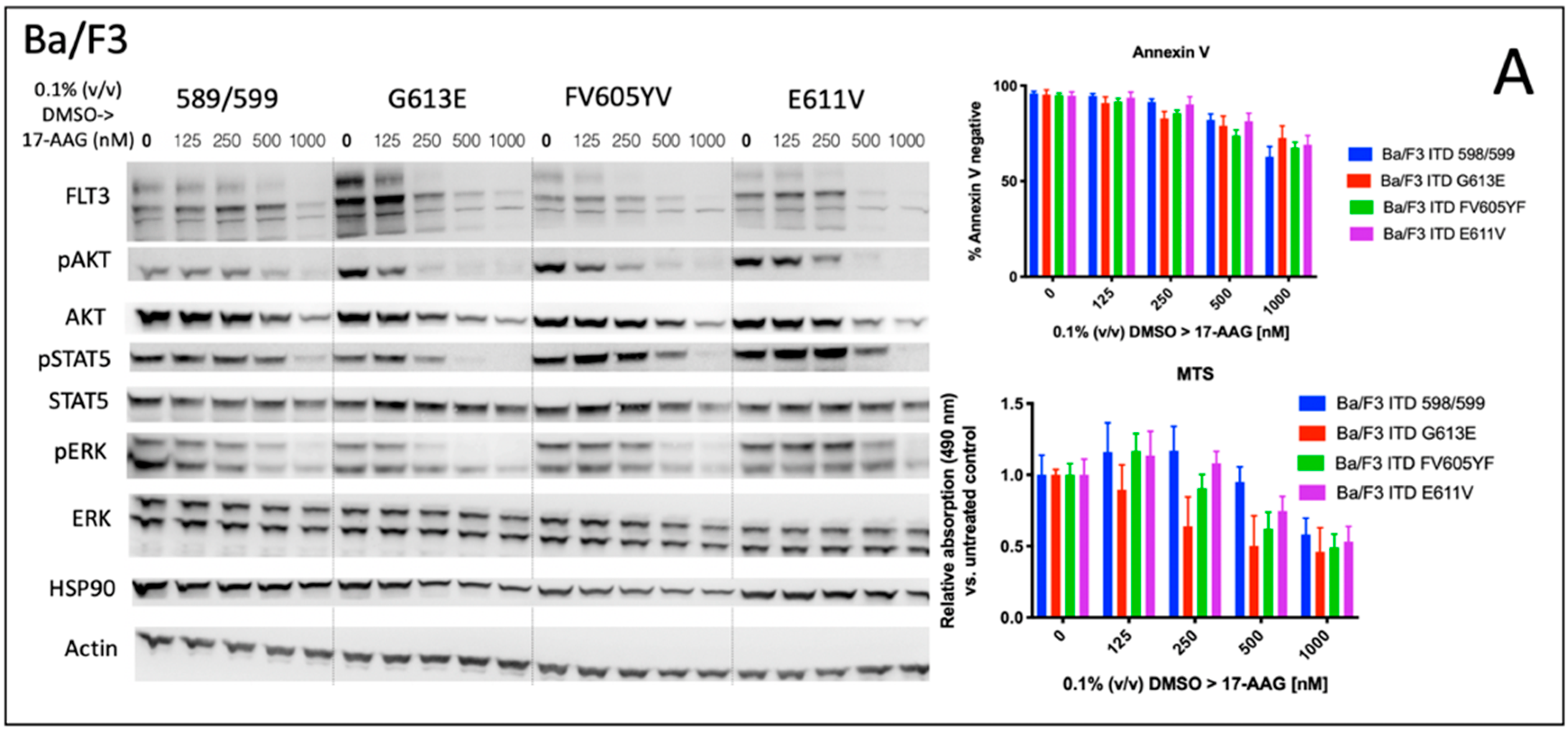
Cells | Free Full-Text | Modulation of FLT3-ITD Localization and Targeting of Distinct Downstream Signaling Pathways as Potential Strategies to Overcome FLT3-Inhibitor Resistance

Detection of FLT3 Internal Tandem Duplication in Targeted, Short-Read-Length, Next-Generation Sequencing Data - ScienceDirect

Tandem duplication PCR: an ultrasensitive assay for the detection of internal tandem duplications of the FLT3 gene. - Abstract - Europe PMC

The FLT3-ITD mutation and the expression of its downstream signaling intermediates STAT5 and Pim-1 are positively correlated with CXCR4 expression in patients with acute myeloid leukemia | Scientific Reports

Figure 4 from Detection of FLT3 (FMS-like Tyrosine Kinase) Internal Tandem Duplication (ITD) Mutation using Next Generation Sequencing Technology and Nested PCR | Semantic Scholar
![PDF] FLT3-ITD, NPM1, and DNMT3A Gene Mutations and Risk Factors in Normal Karyotype Acute Myeloid Leukemia and Myelodysplastic Syndrome Patients in Upper Northern Thailand | Semantic Scholar PDF] FLT3-ITD, NPM1, and DNMT3A Gene Mutations and Risk Factors in Normal Karyotype Acute Myeloid Leukemia and Myelodysplastic Syndrome Patients in Upper Northern Thailand | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/c42682134066e9ecb7be4d79b2d585c516877b2d/3-Figure1-1.png)
PDF] FLT3-ITD, NPM1, and DNMT3A Gene Mutations and Risk Factors in Normal Karyotype Acute Myeloid Leukemia and Myelodysplastic Syndrome Patients in Upper Northern Thailand | Semantic Scholar
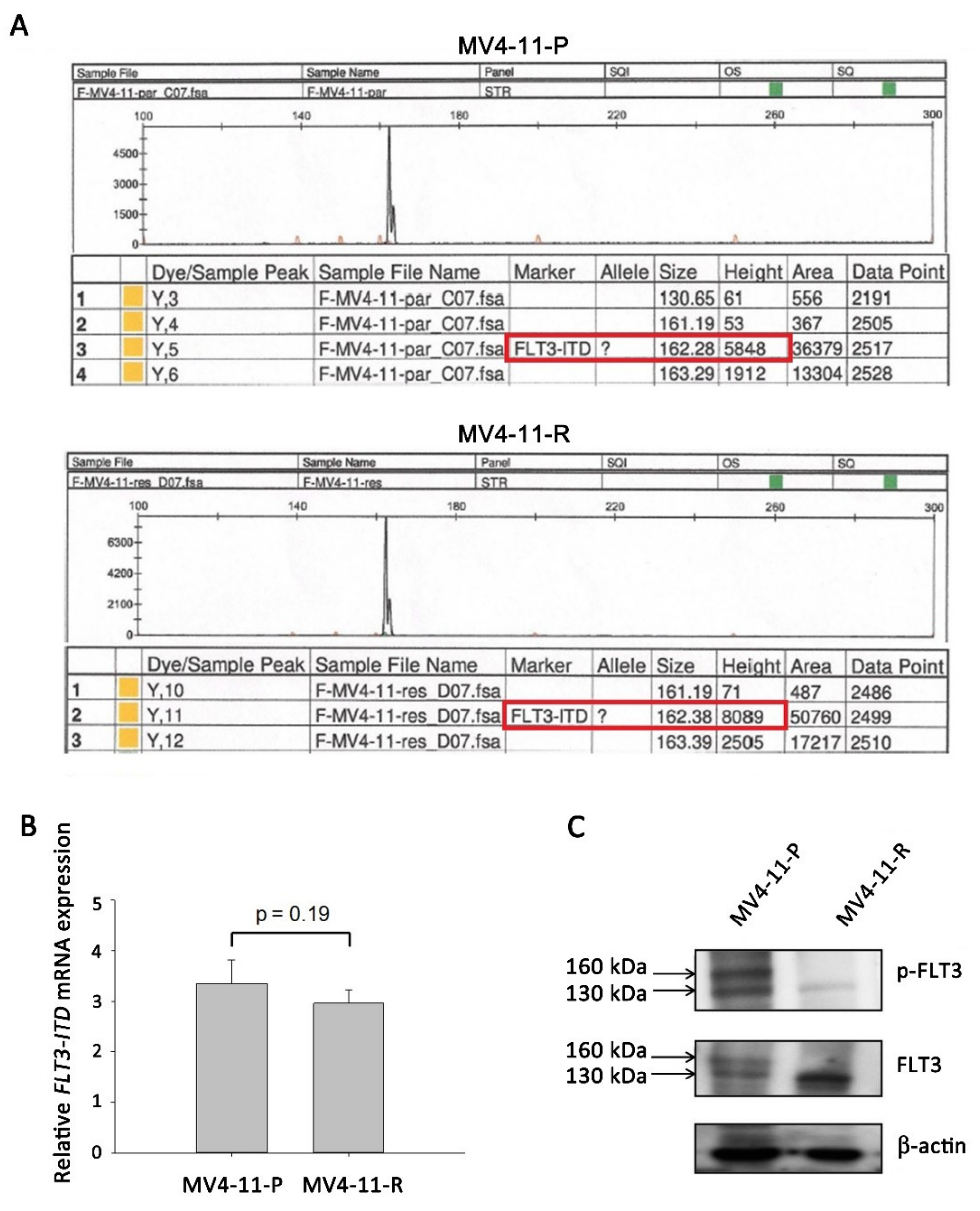
IJMS | Free Full-Text | Cytarabine-Resistant FLT3-ITD Leukemia Cells are Associated with TP53 Mutation and Multiple Pathway Alterations—Possible Therapeutic Efficacy of Cabozantinib




