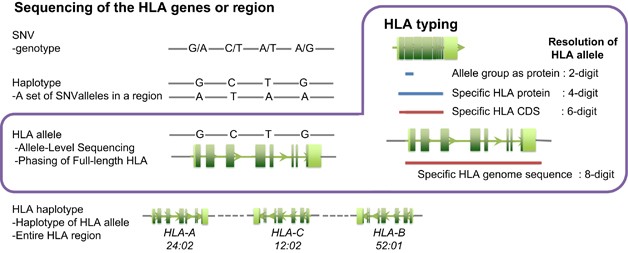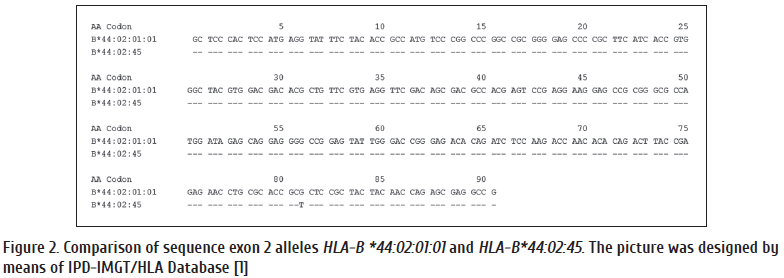
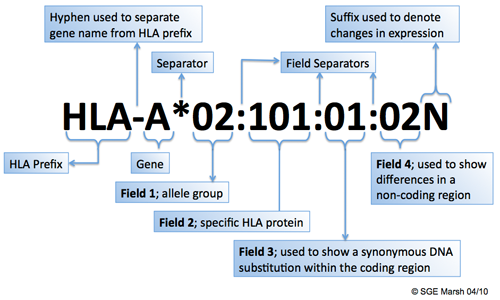
Identification of the new HLA-B*44:02:45, DQB1*02:85, DQB1*06:210, DRB1*01:01:30 alleles by monoallelic Sanger sequencing
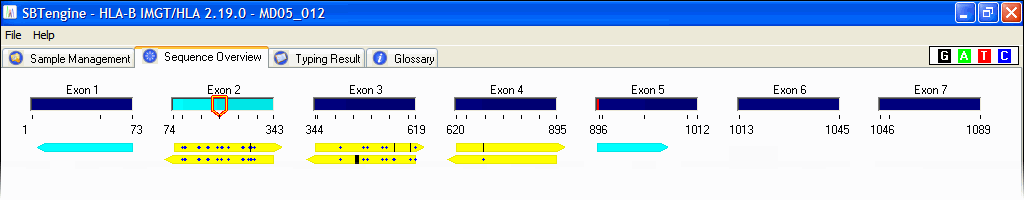
SOAPTyping: an open-source and cross-platform tool for sequence-based typing for HLA class I and II alleles
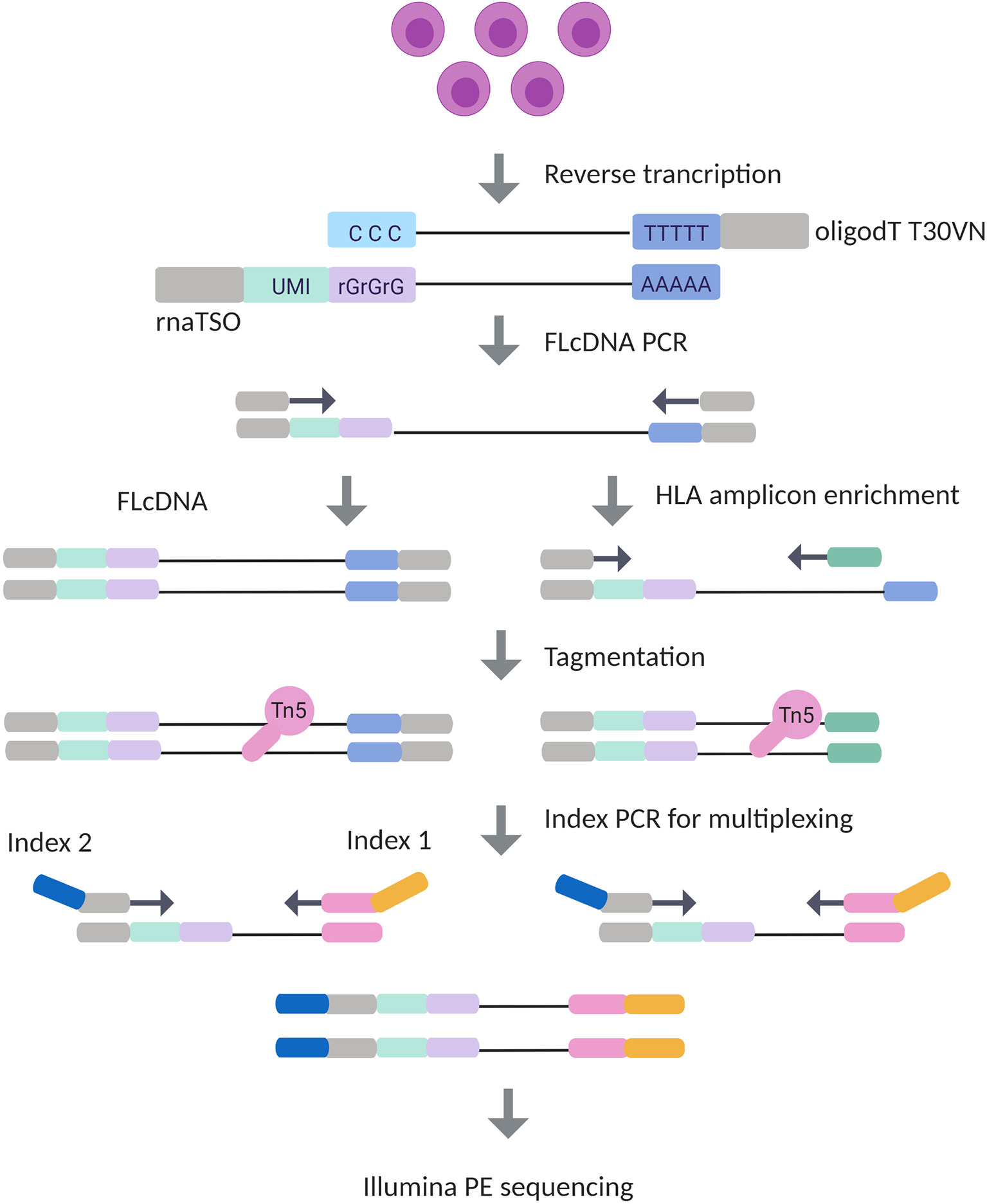
Frontiers | HLA RNA Sequencing With Unique Molecular Identifiers Reveals High Allele-Specific Variability in mRNA Expression
Vitamin D Responsive Elements within the HLA-DRB1 Promoter Region in Sardinian Multiple Sclerosis Associated Alleles | PLOS ONE

Personalized HLA typing leads to the discovery of novel HLA alleles and tumor‐specific HLA variants - Anzar - 2022 - HLA - Wiley Online Library
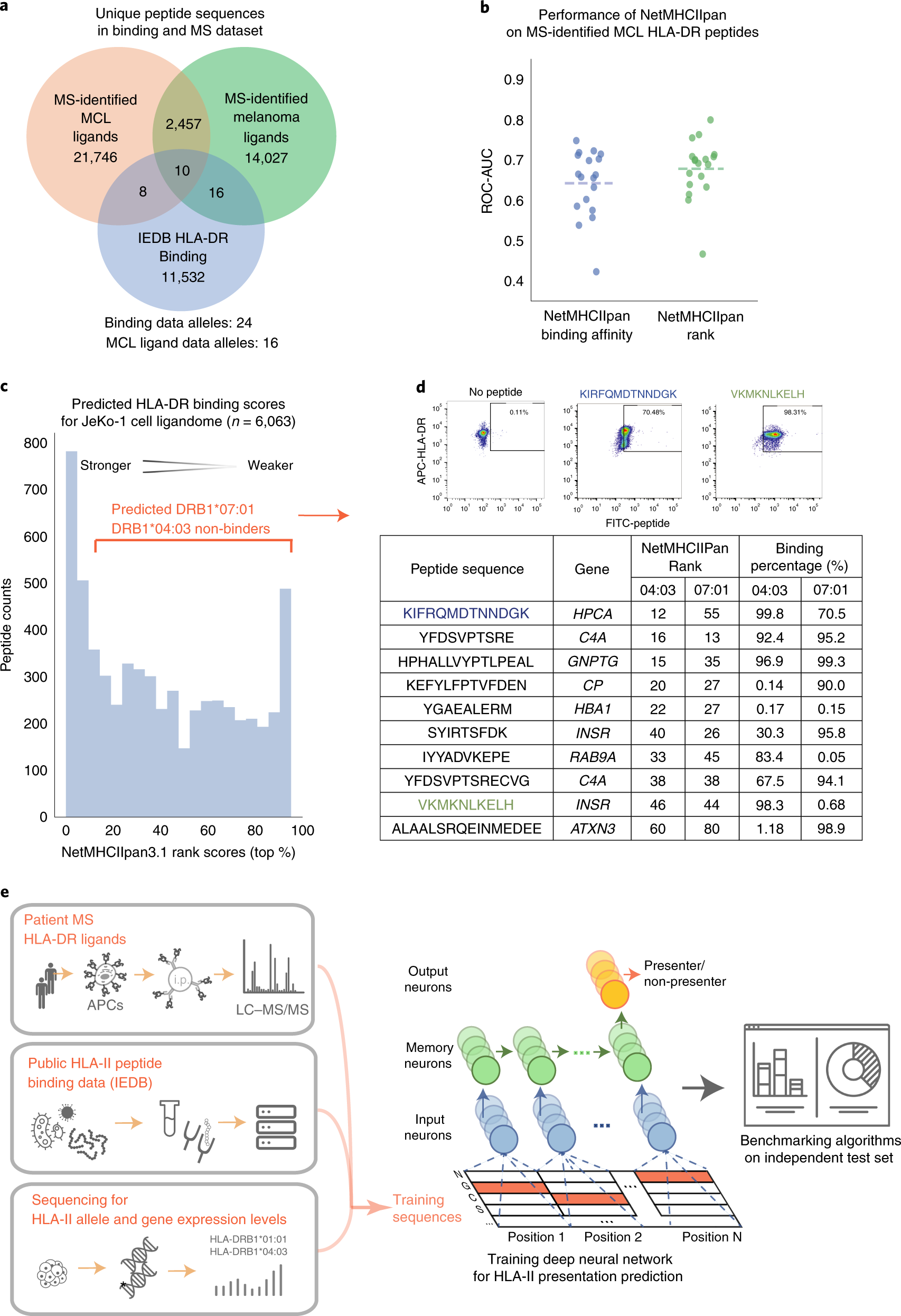
Predicting HLA class II antigen presentation through integrated deep learning | Nature Biotechnology

Frontiers | Unsupervised Mining of HLA-I Peptidomes Reveals New Binding Motifs and Potential False Positives in the Community Database

Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility - ScienceDirect
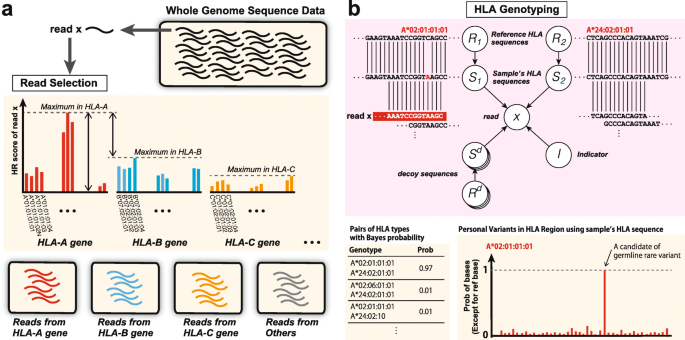
ALPHLARD: a Bayesian method for analyzing HLA genes from whole genome sequence data | BMC Genomics | Full Text

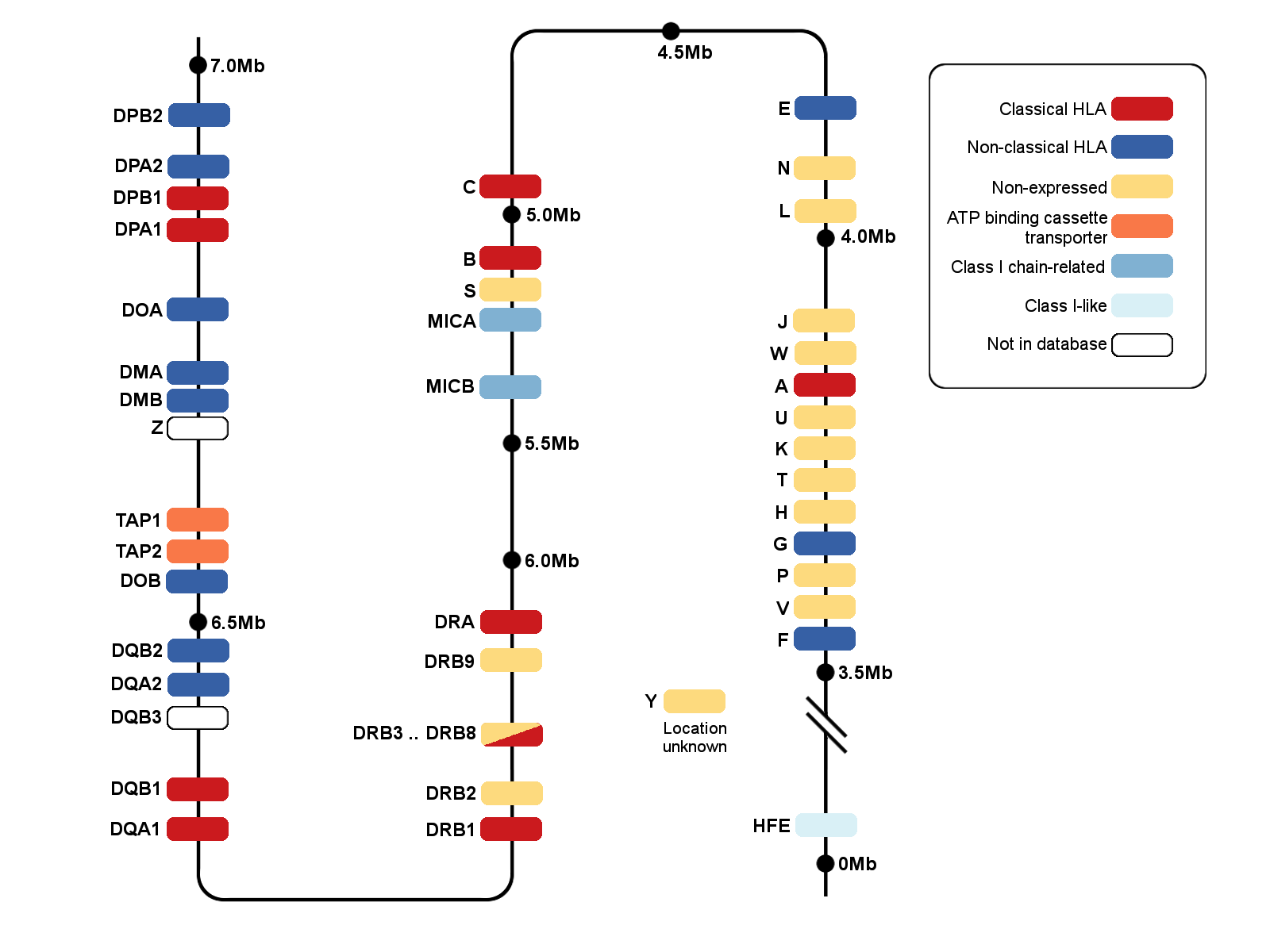
Alignment formats available from the IMGT/HLA and IMGT/MHC Database.... | Download Scientific Diagram



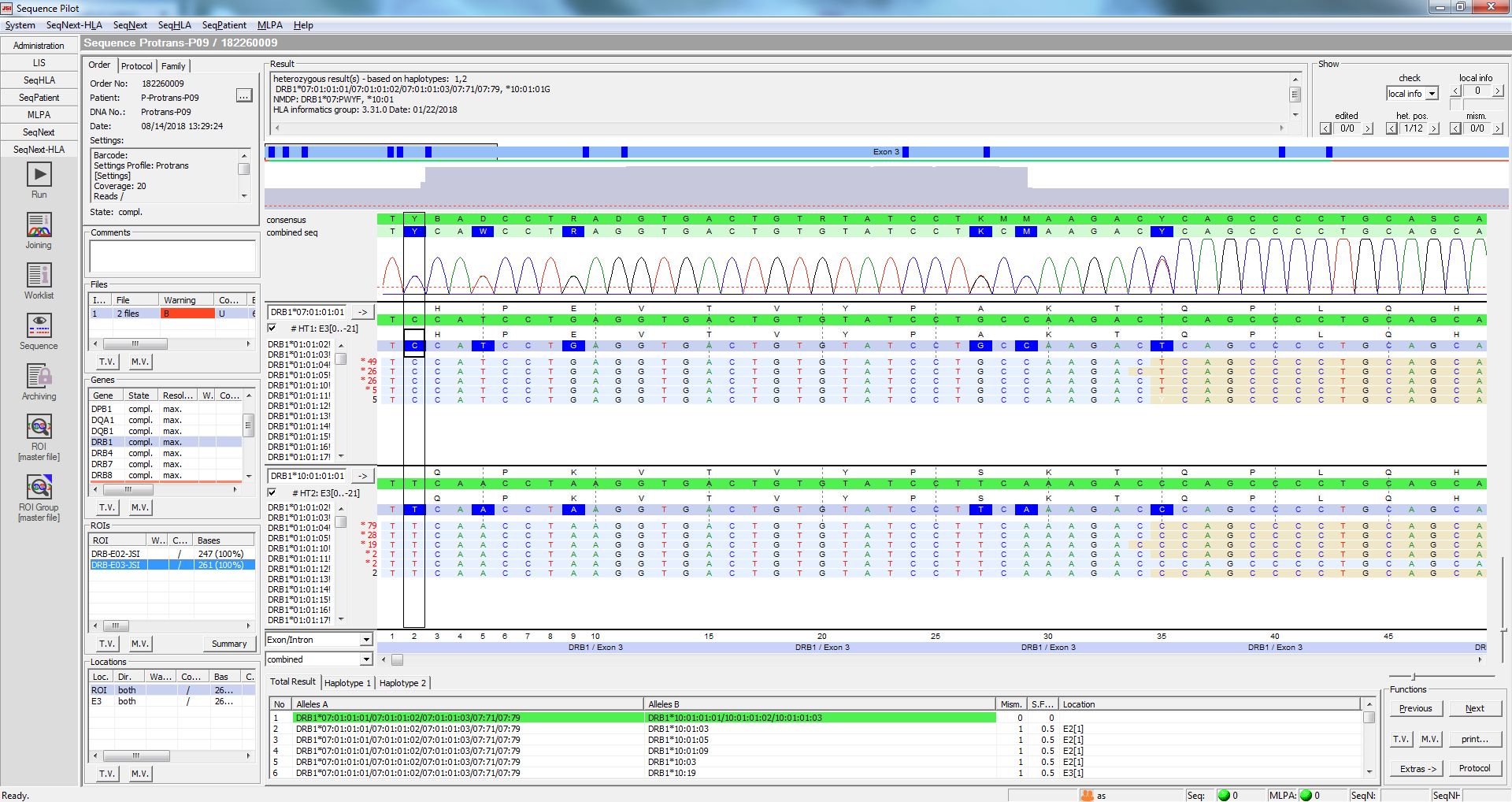

![2020-09-01] IIBMP2020 Generating annotation texts of HLA sequences with antigen classes by a T5 2020-09-01] IIBMP2020 Generating annotation texts of HLA sequences with antigen classes by a T5](https://image.slidesharecdn.com/ekiibmp2020fin-200901093859/85/20200901-iibmp2020-generating-annotation-texts-of-hla-sequences-with-antigen-classes-by-a-t5-model-using-insdc-1-320.jpg?cb=1668360411)