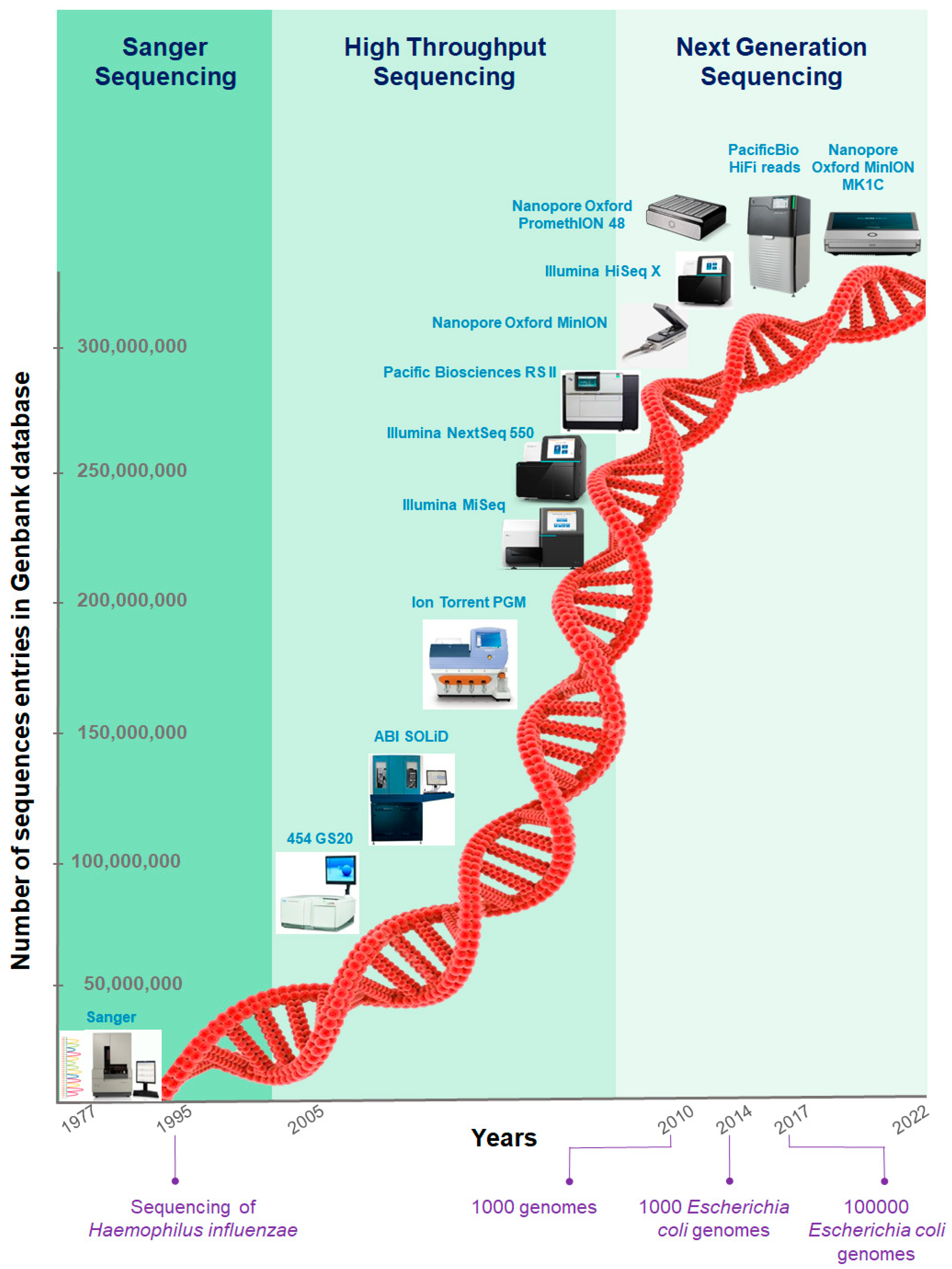
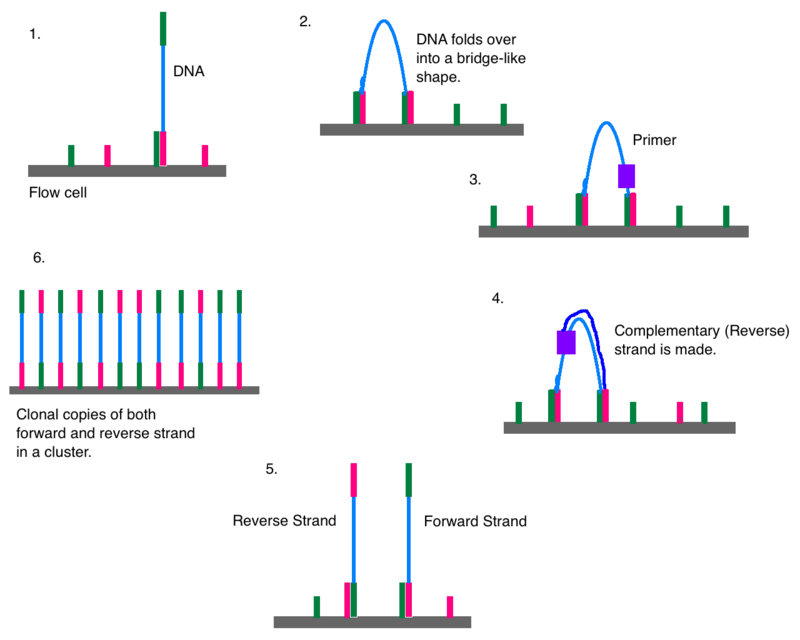
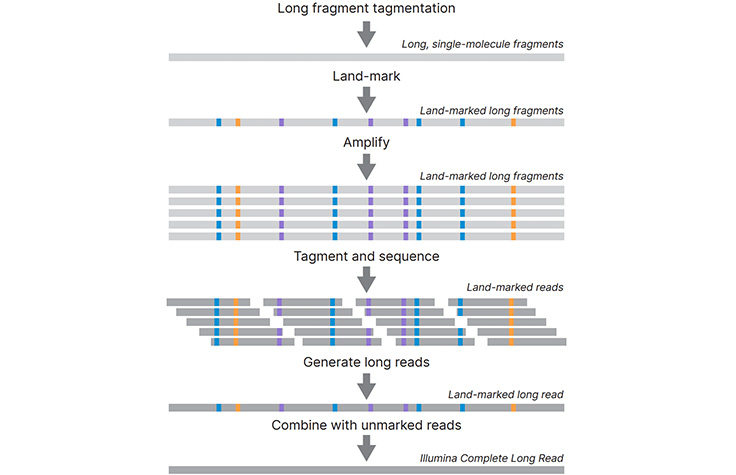
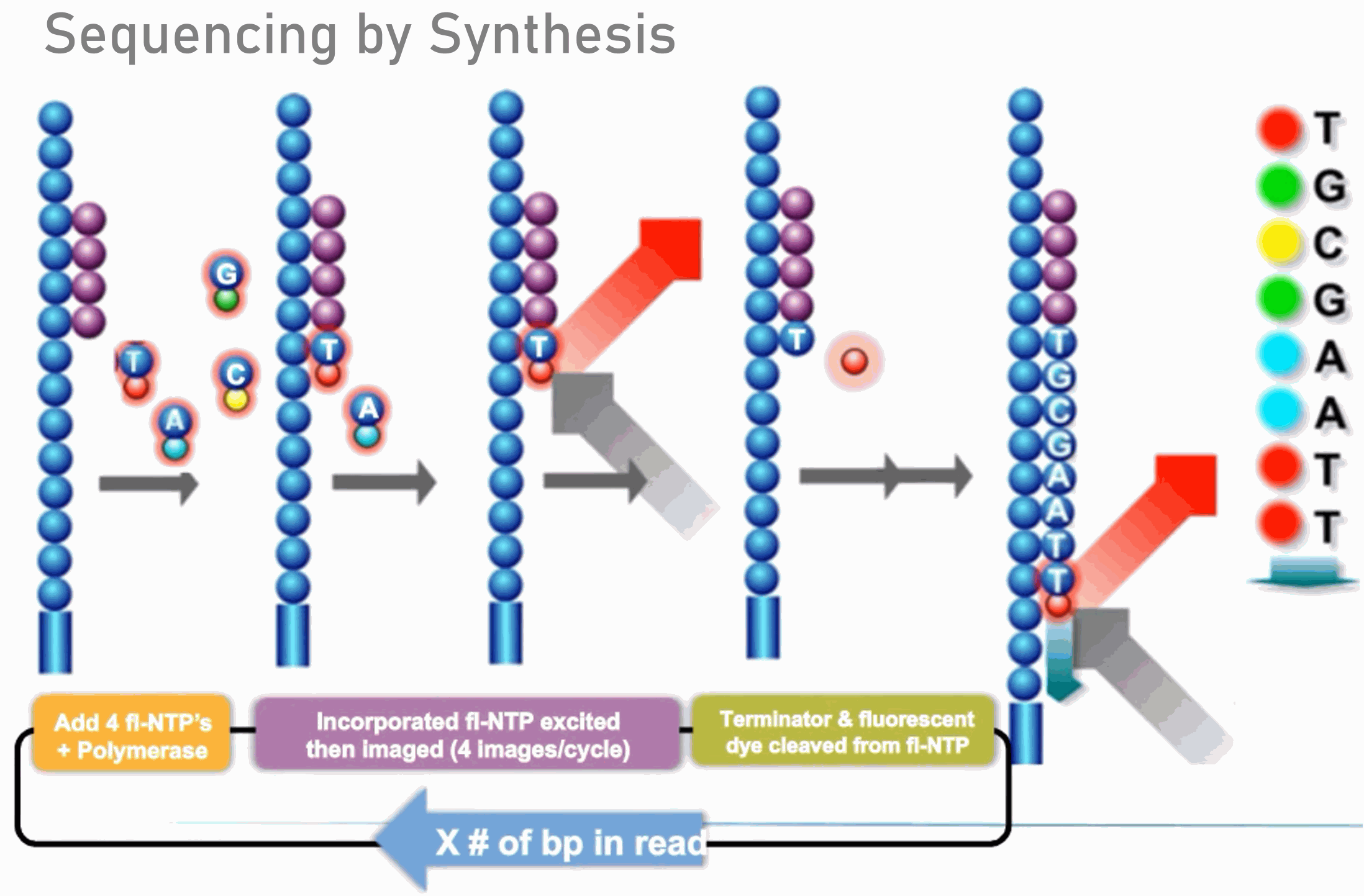
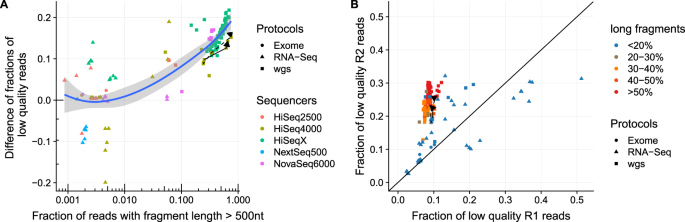
Histogram of read variants' lengths in Illumina sequencing data. The... | Download Scientific Diagram
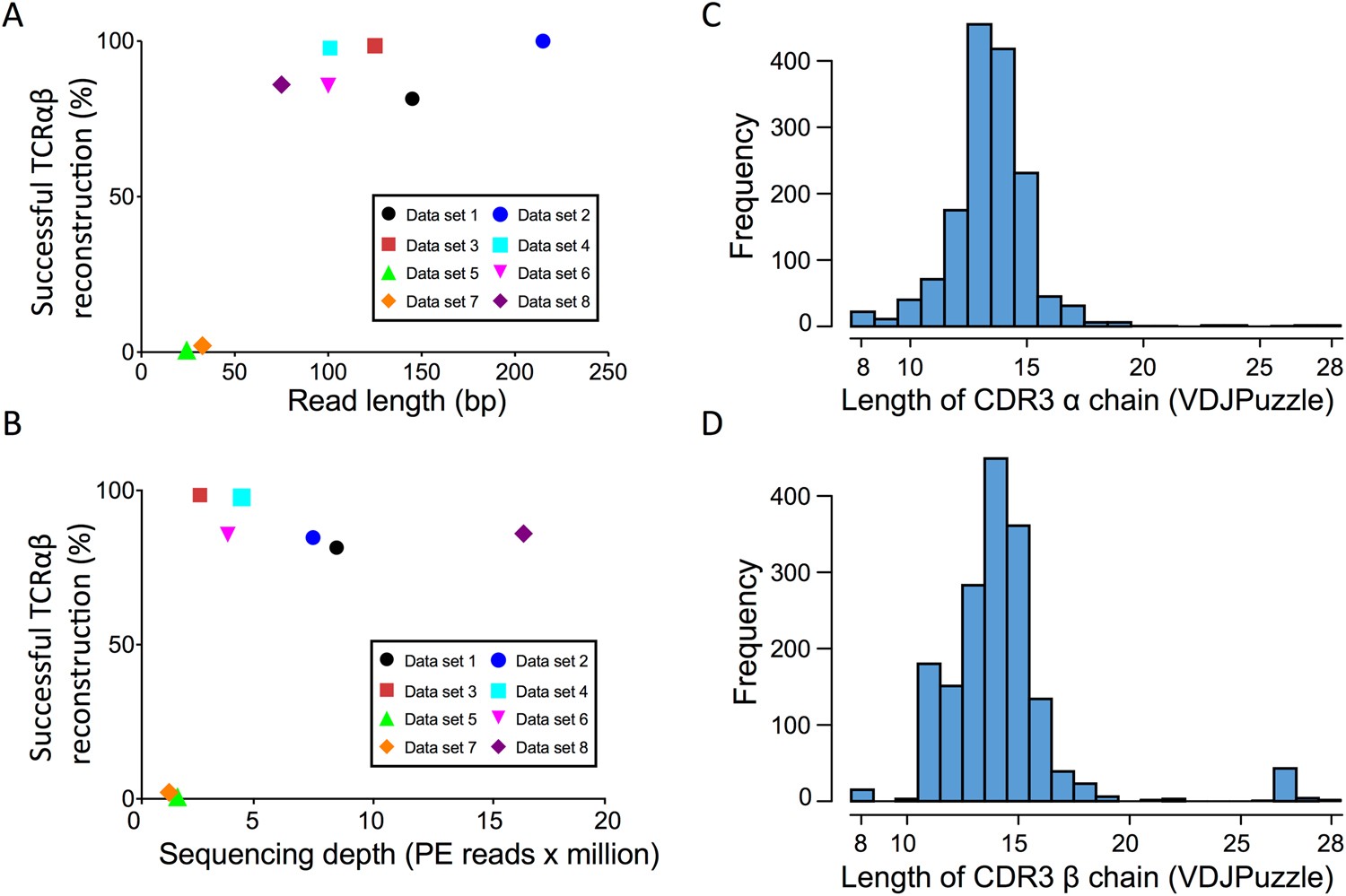
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

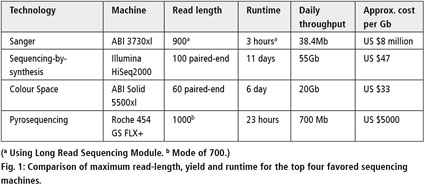
Illumina HiSeq v4 Sequencing Services Yielding 1Tb Data / Week Now Available on Genohub | Genohub Blog

The distribution of read lengths in 454, Illumina and PacBio budgerigar... | Download Scientific Diagram

Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge