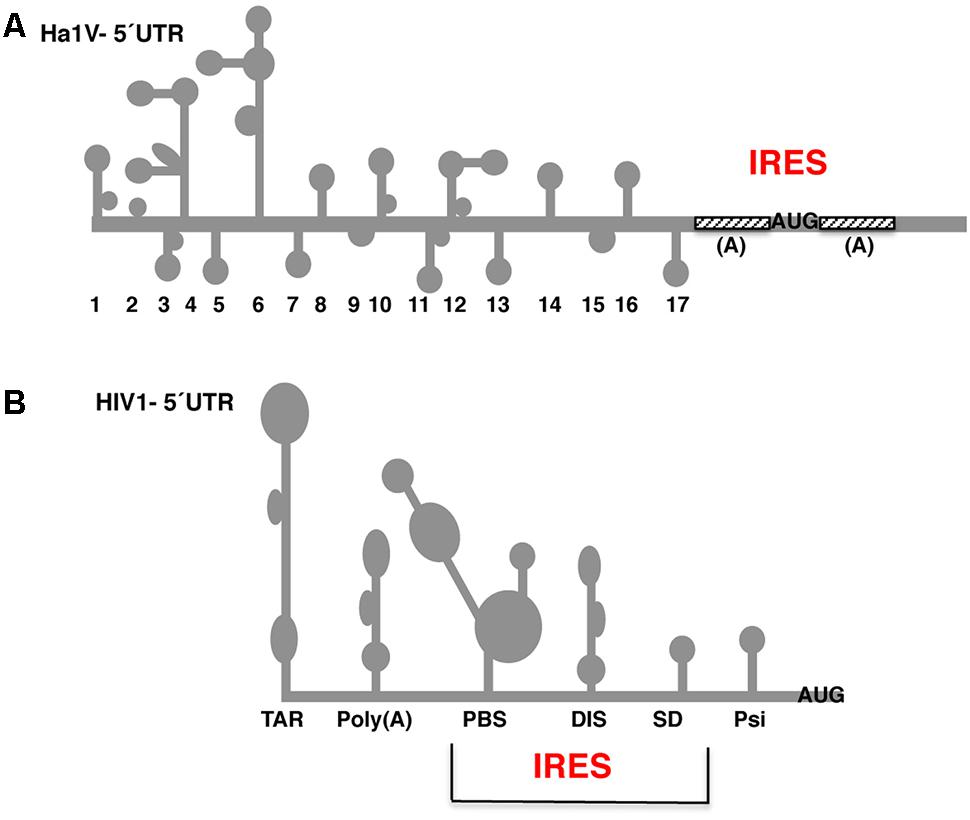
Identification of an IRES within the coding region of the structural protein of human rhinovirus 16 - Shi - 2022 - Journal of Medical Virology - Wiley Online Library

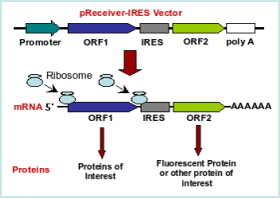
IRES-Dependent Second Gene Expression Is Significantly Lower Than Cap-Dependent First Gene Expression in a Bicistronic Vector: Molecular Therapy
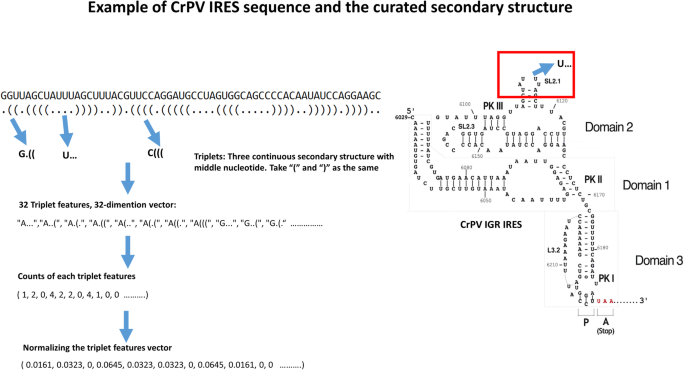
IRESpy: an XGBoost model for prediction of internal ribosome entry sites | BMC Bioinformatics | Full Text
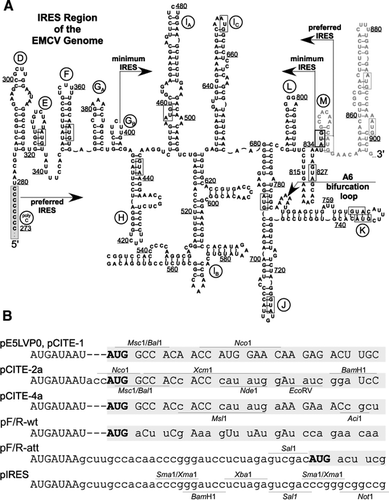
Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location | BioTechniques

Modifying inter-cistronic sequence significantly enhances IRES dependent second gene expression in bicistronic vector: Construction of optimised cassette for gene therapy of familial hypercholesterolemia - ScienceDirect

A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity | PNAS
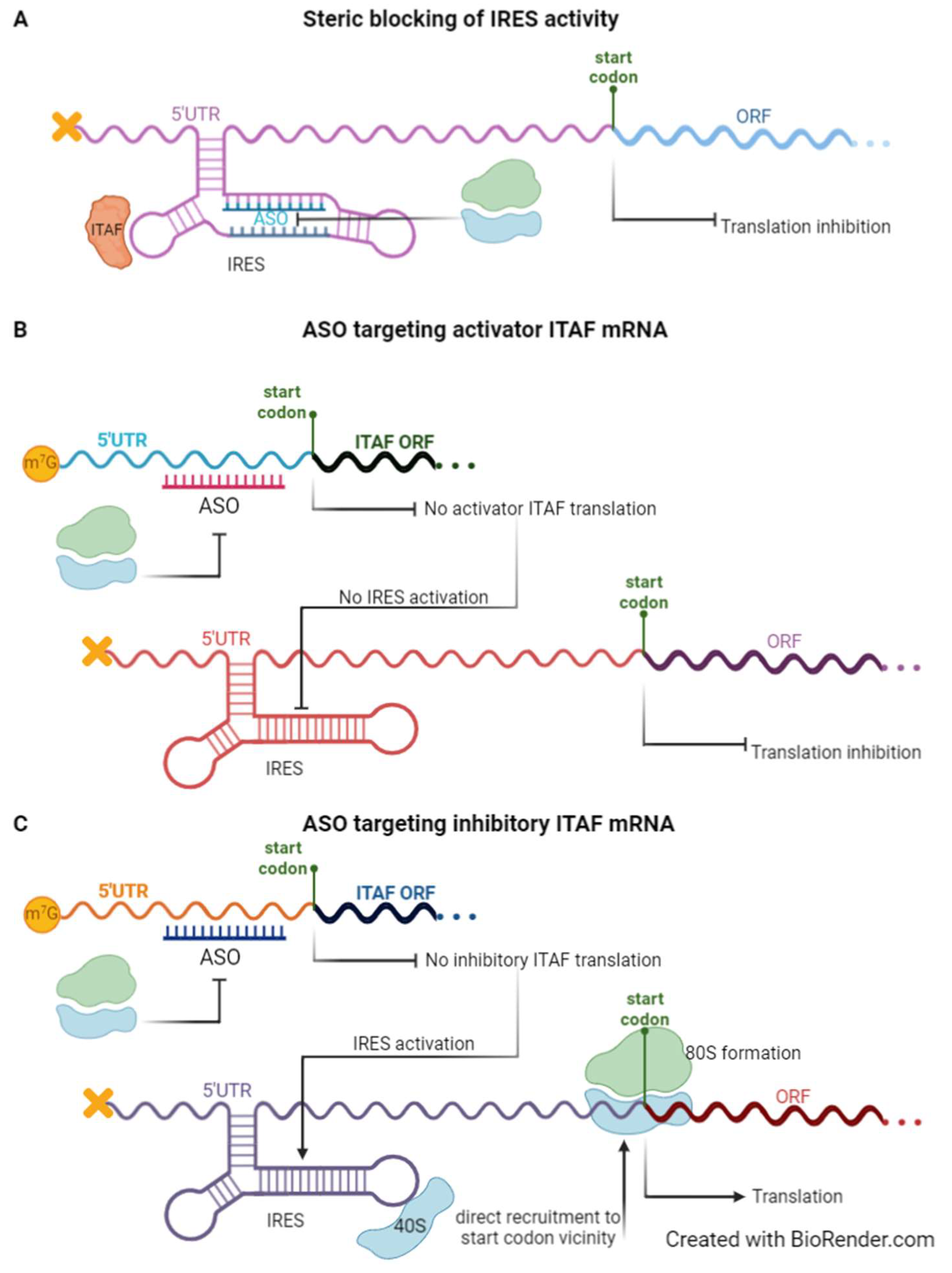
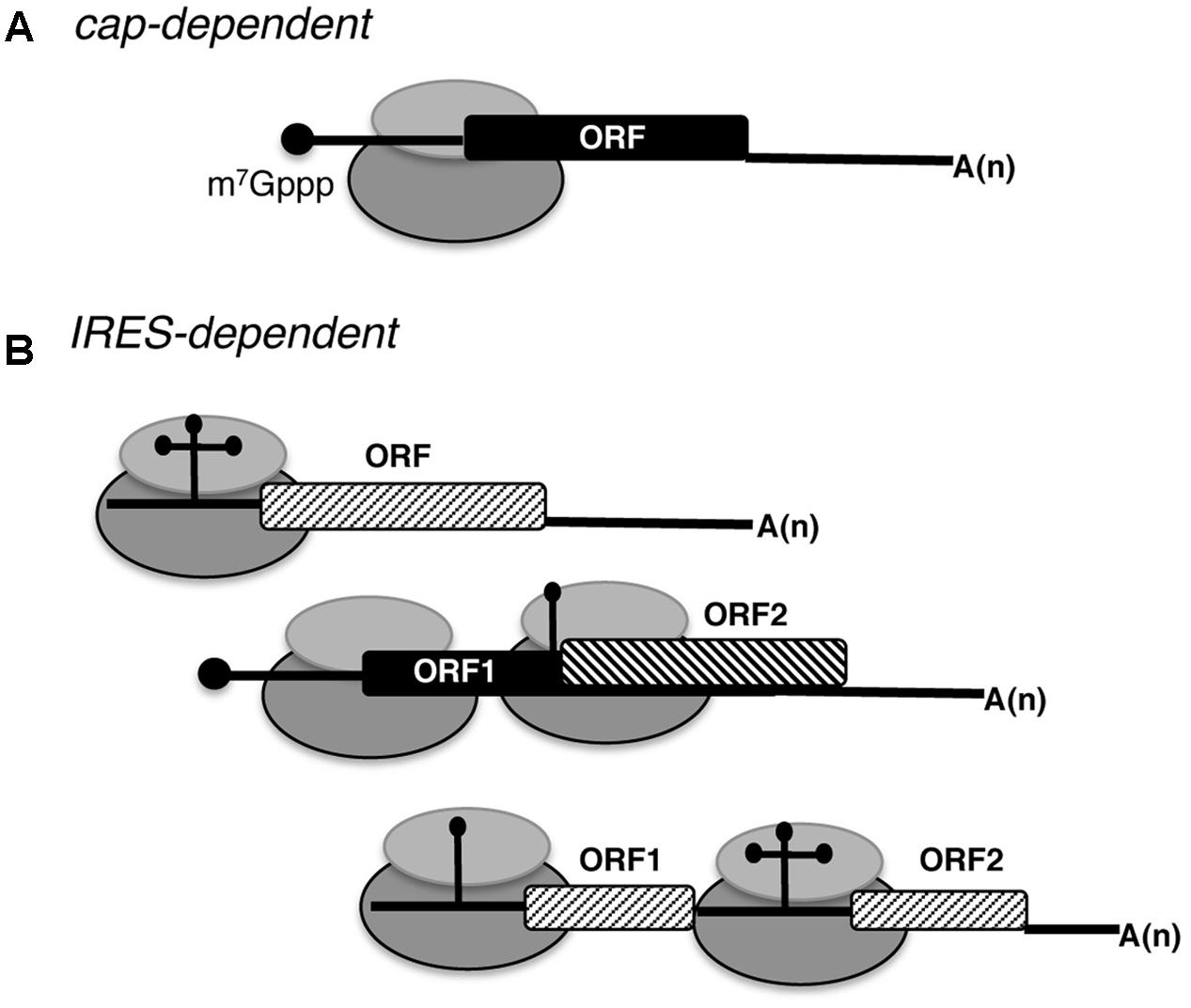
Biomedicines | Free Full-Text | Internal Ribosome Entry Site (IRES)-Mediated Translation and Its Potential for Novel mRNA-Based Therapy Development

Internal Ribosome Entry Site-mediated Translation of a Mammalian mRNA Is Regulated by Amino Acid Availability* - Journal of Biological Chemistry

The IRES element type I. Each of the sequence motifs characteristic for... | Download Scientific Diagram

IRESbase: a Comprehensive Database of Experimentally Validated Internal Ribosome Entry Sites | bioRxiv