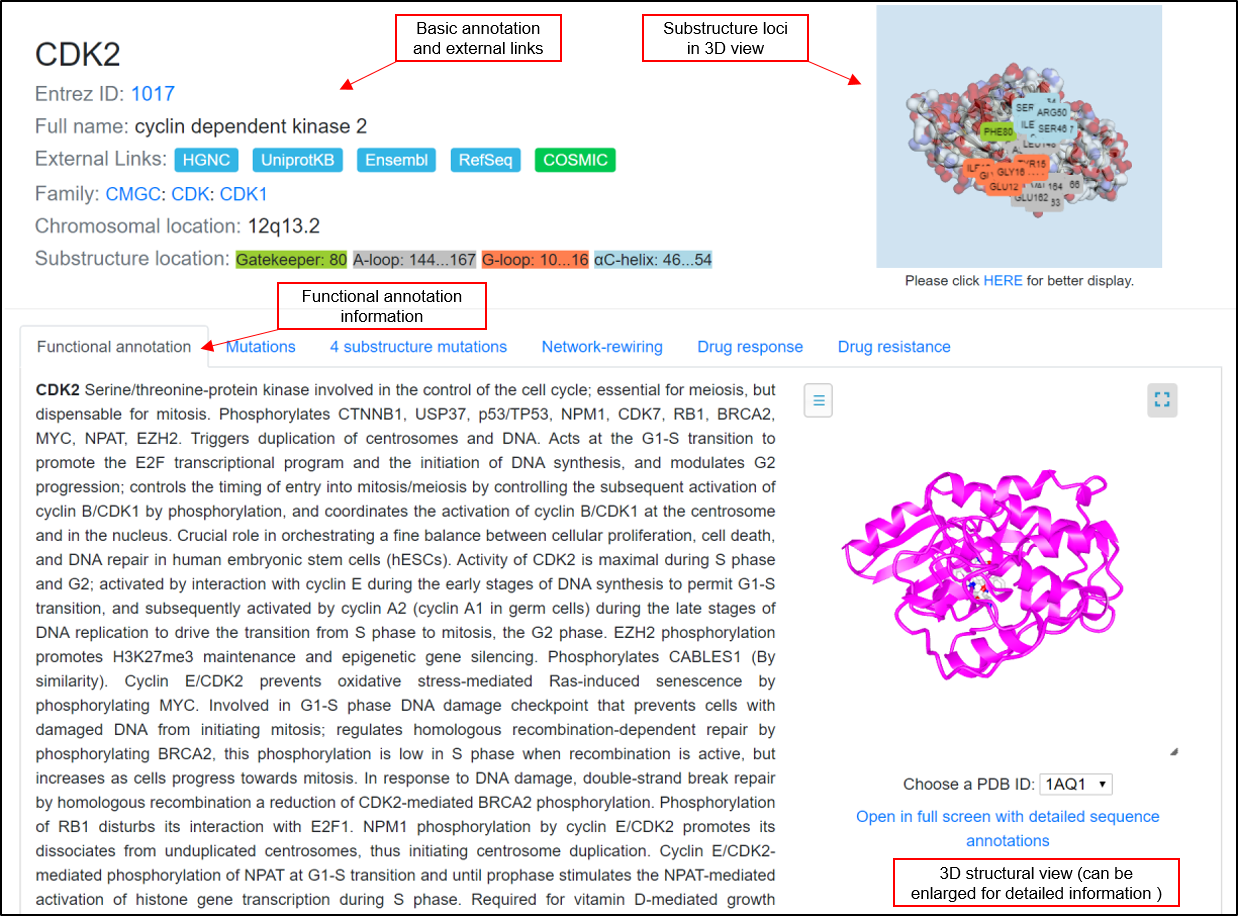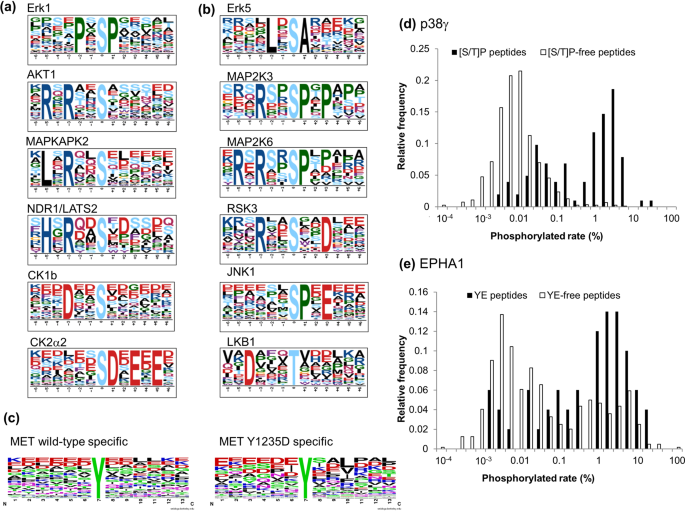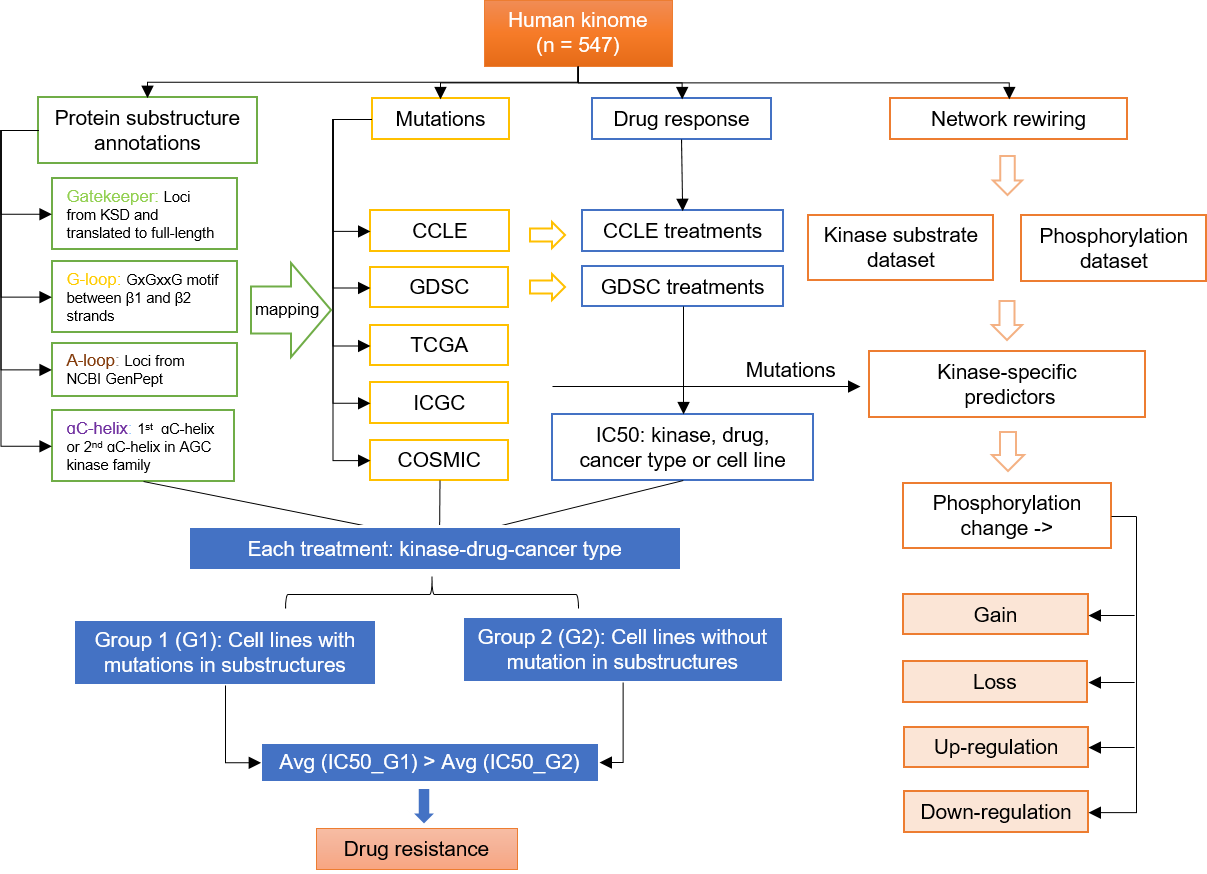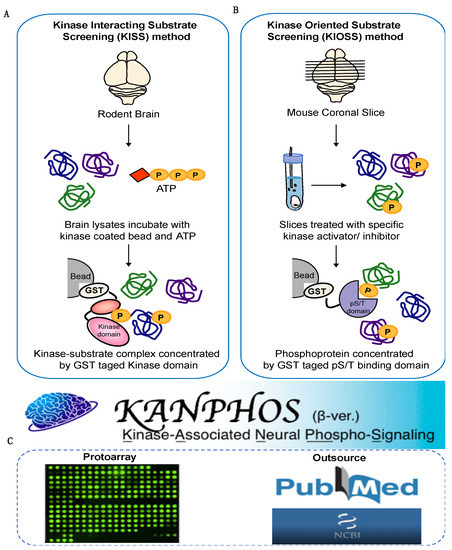
Cells | Free Full-Text | KANPHOS: A Database of Kinase-Associated Neural Protein Phosphorylation in the Brain
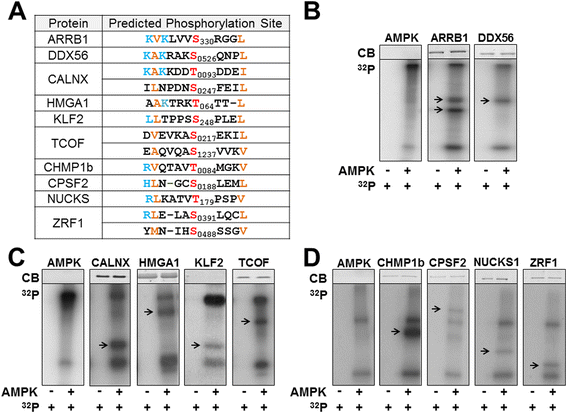
Identification of AMP-activated protein kinase targets by a consensus sequence search of the proteome | BMC Systems Biology | Full Text

PhosphoPredict: A bioinformatics tool for prediction of human kinase-specific phosphorylation substrates and sites by integrating heterogeneous feature selection | Scientific Reports

Kinase consensus sequences. Consensus sequences of some of the kinases... | Download Scientific Diagram

The consensus sequences of Protein kinase domain (Pkinase, pfam00069)... | Download Scientific Diagram

Identification of Phosphorylation Consensus Sequences and Endogenous Neuronal Substrates of the Psychiatric Risk Kinase TNIK | Journal of Pharmacology and Experimental Therapeutics

A Structurally-Validated Multiple Sequence Alignment of 497 Human Protein Kinase Domains | Scientific Reports
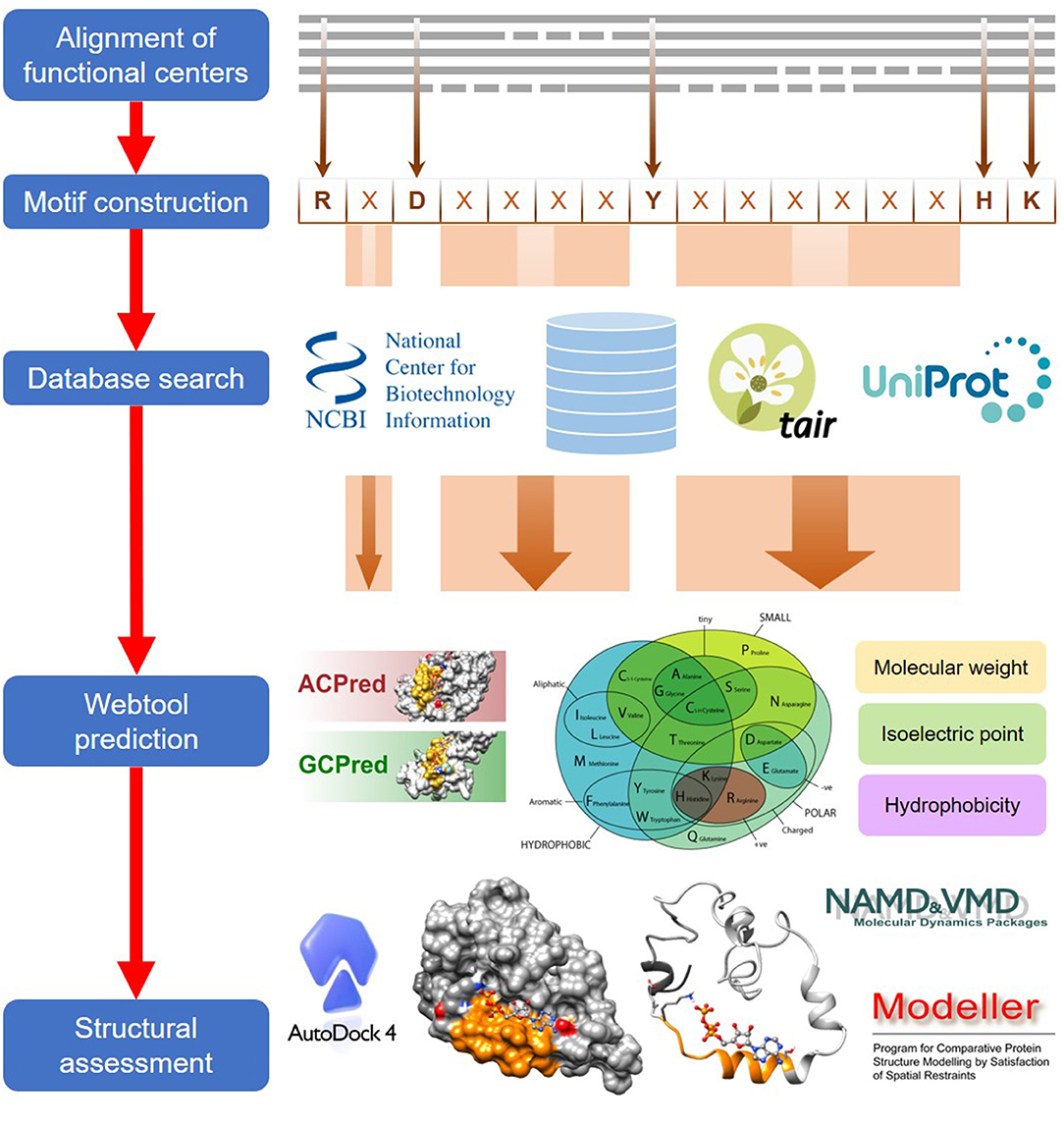
Frontiers | Computational Identification of Functional Centers in Complex Proteins: A Step-by-Step Guide With Examples
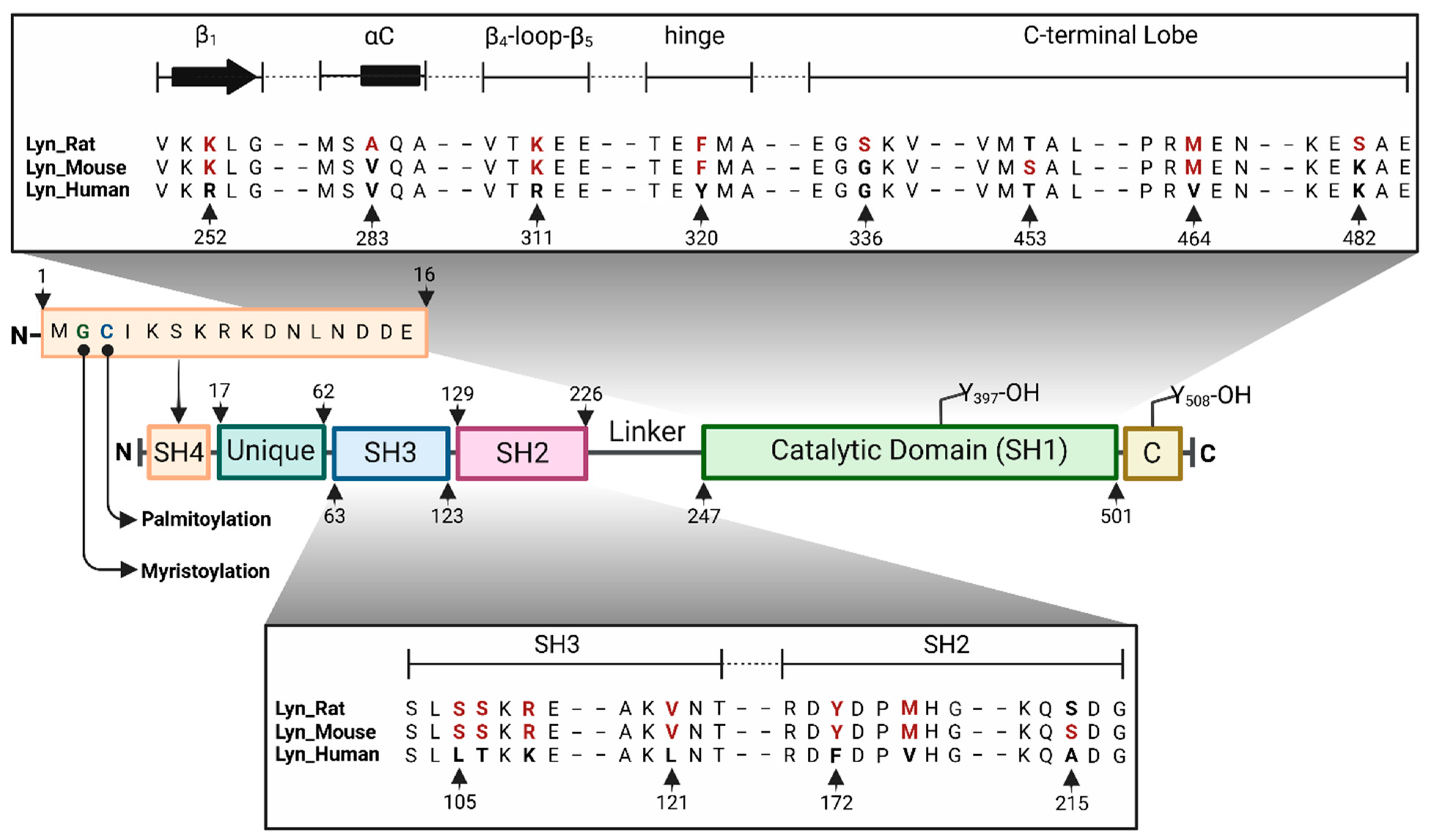
Kinases and Phosphatases | Free Full-Text | Lyn Kinase Structure, Regulation, and Involvement in Neurodegenerative Diseases: A Mini Review

Kinase consensus sequences. Consensus sequences of some of the kinases... | Download Scientific Diagram

Life | Free Full-Text | Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases

An illustration of extraction of different consensus sequence motifs by... | Download Scientific Diagram

A rapid computational approach identifies SPICE1 as an Aurora kinase substrate | Molecular Biology of the Cell
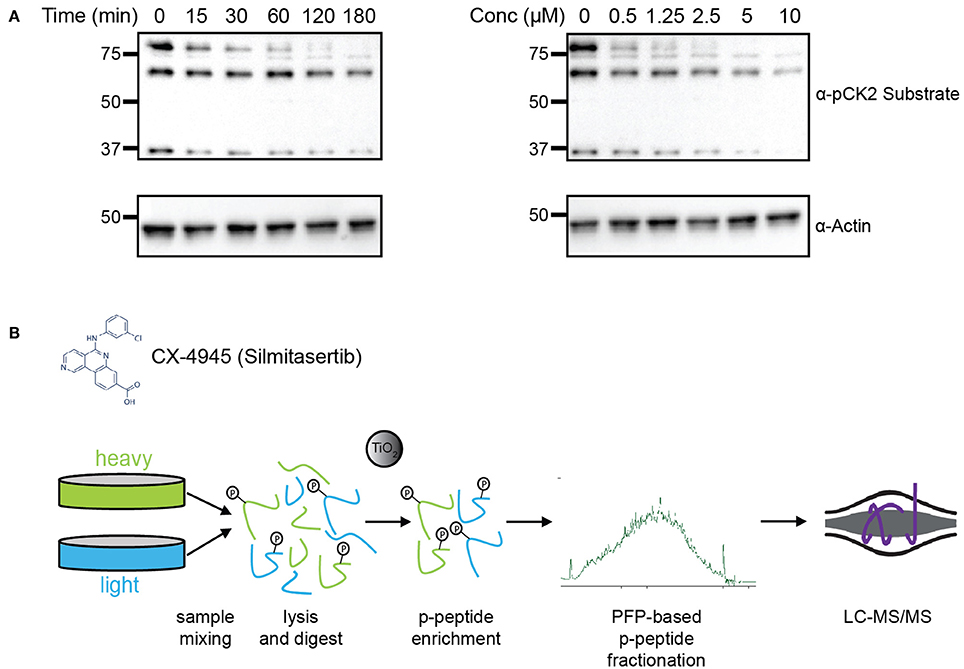
Frontiers | Identification of Candidate Casein Kinase 2 Substrates in Mitosis by Quantitative Phosphoproteomics

Interrogating Kinase–Substrate Relationships with Proximity Labeling and Phosphorylation Enrichment | Journal of Proteome Research

Substrate Recognition Mechanism of Atypical Protein Kinase Cs Revealed by the Structure of PKCι in Complex with a Substrate Peptide from Par-3 - ScienceDirect
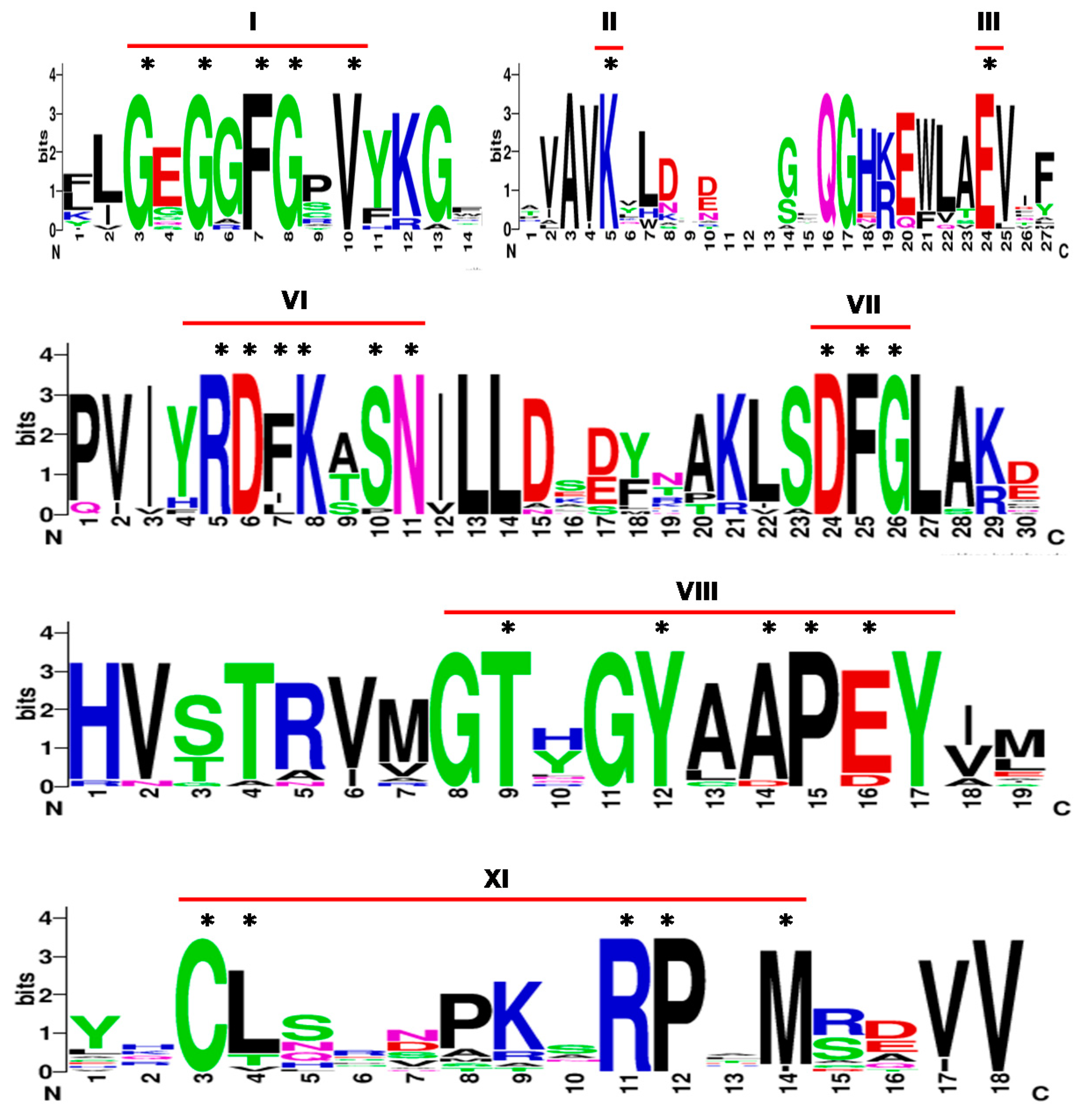
Plants | Free Full-Text | Characterization of Atypical Protein Tyrosine Kinase (PTK) Genes and Their Role in Abiotic Stress Response in Rice

Mapping and analysis of phosphorylation sites: a quick guide for cell biologists | Molecular Biology of the Cell