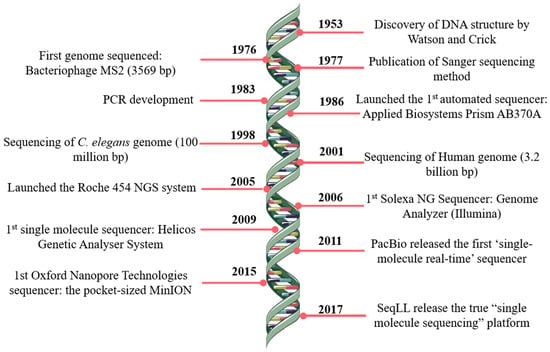
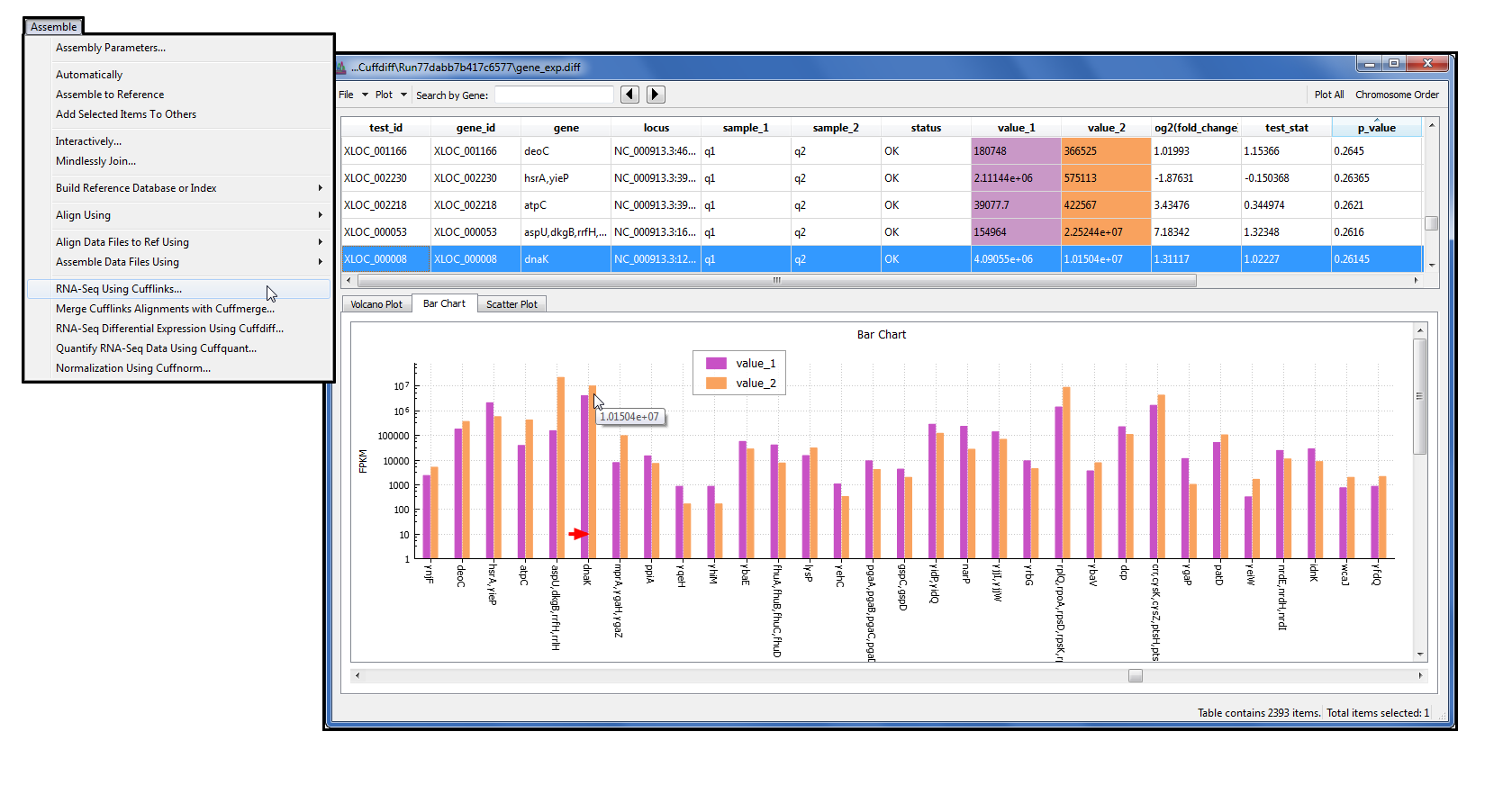
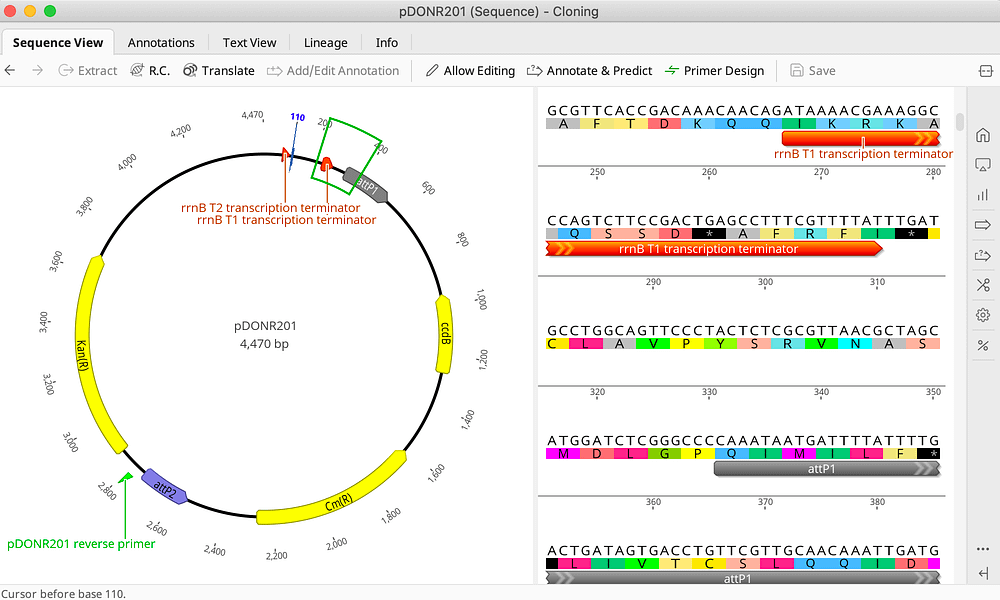
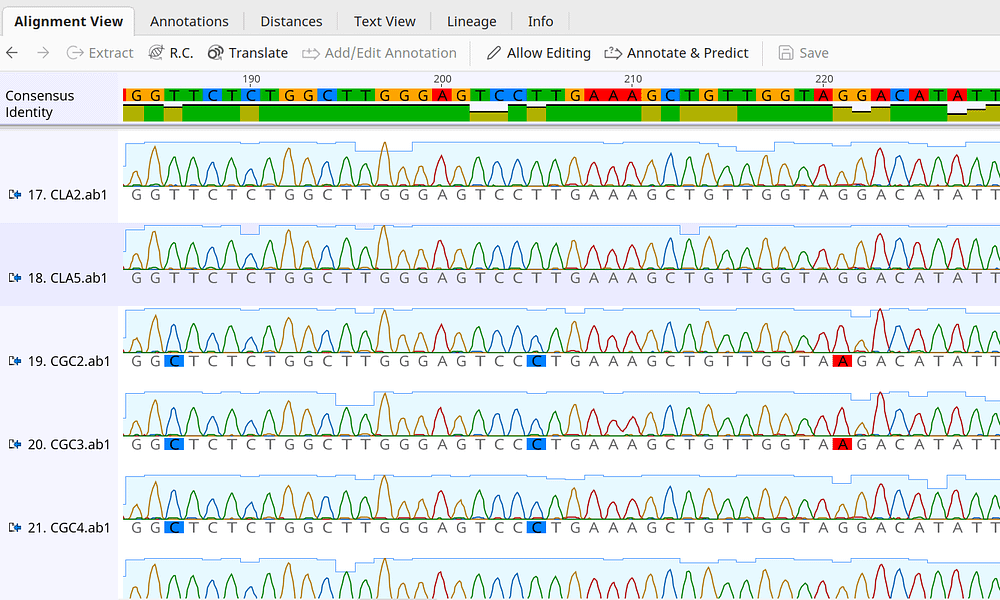
Analysis of Mixed Sequencing Chromatograms and Its Application in Direct 16S rRNA Gene Sequencing of Polymicrobial Samples | Journal of Clinical Microbiology

Low-Cost, User-Friendly, All-Integrated Smartphone-Based Microplate Reader for Optical-Based Biological and Chemical Analyses | Analytical Chemistry
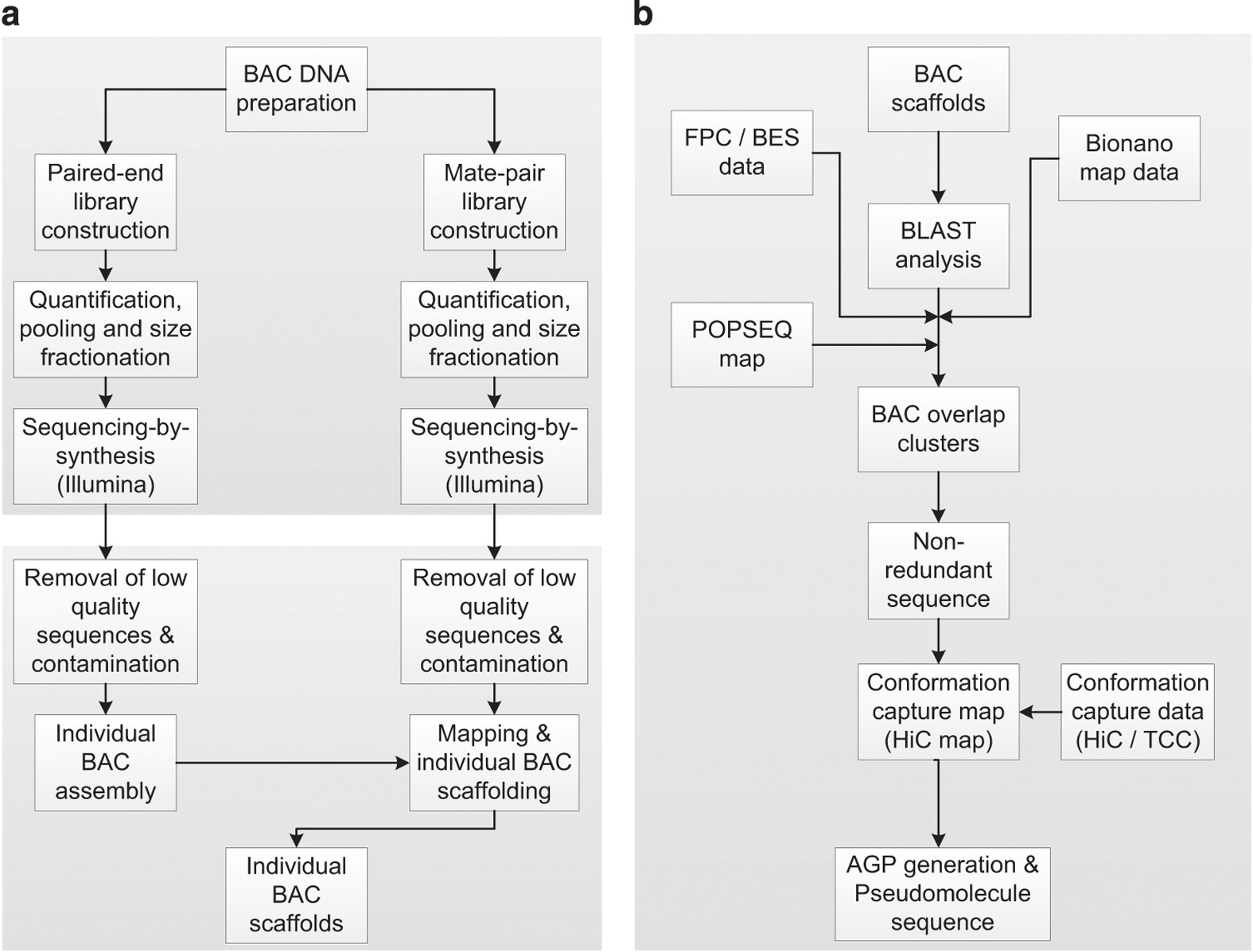
Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. | Scientific Data
![PDF] Mixed Sequence Reader: A Program for Analyzing DNA Sequences with Heterozygous Base Calling | Semantic Scholar PDF] Mixed Sequence Reader: A Program for Analyzing DNA Sequences with Heterozygous Base Calling | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/957a53c4cb6cfbc76c655cef9acec295b0123949/4-Figure2-1.png)
PDF] Mixed Sequence Reader: A Program for Analyzing DNA Sequences with Heterozygous Base Calling | Semantic Scholar
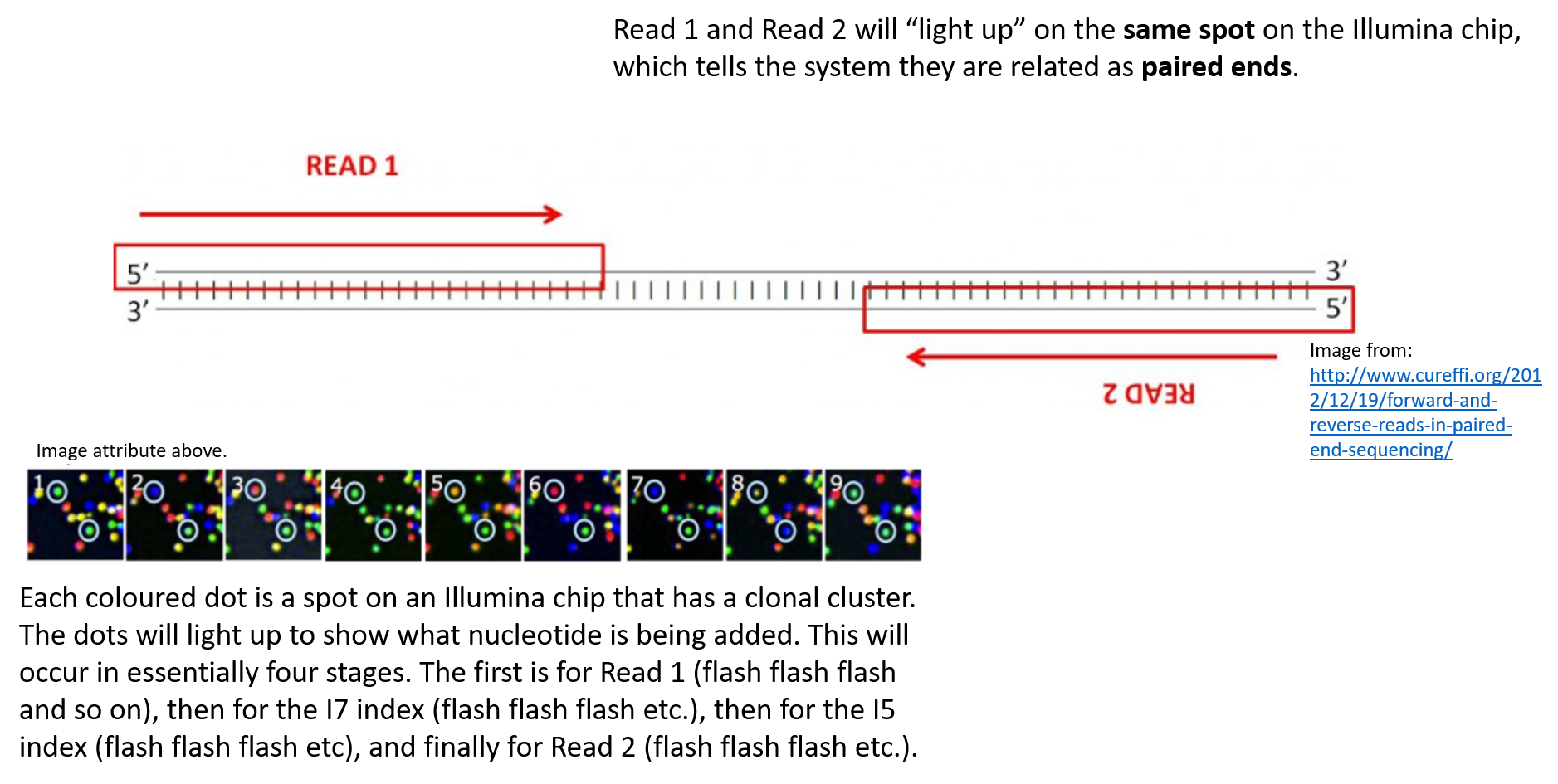
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics
Multi-amplicon microbiome data analysis pipelines for mixed orientation sequences using QIIME2: Assessing reference database, variable region and pre-processing bias in classification of mock bacterial community samples | PLOS ONE
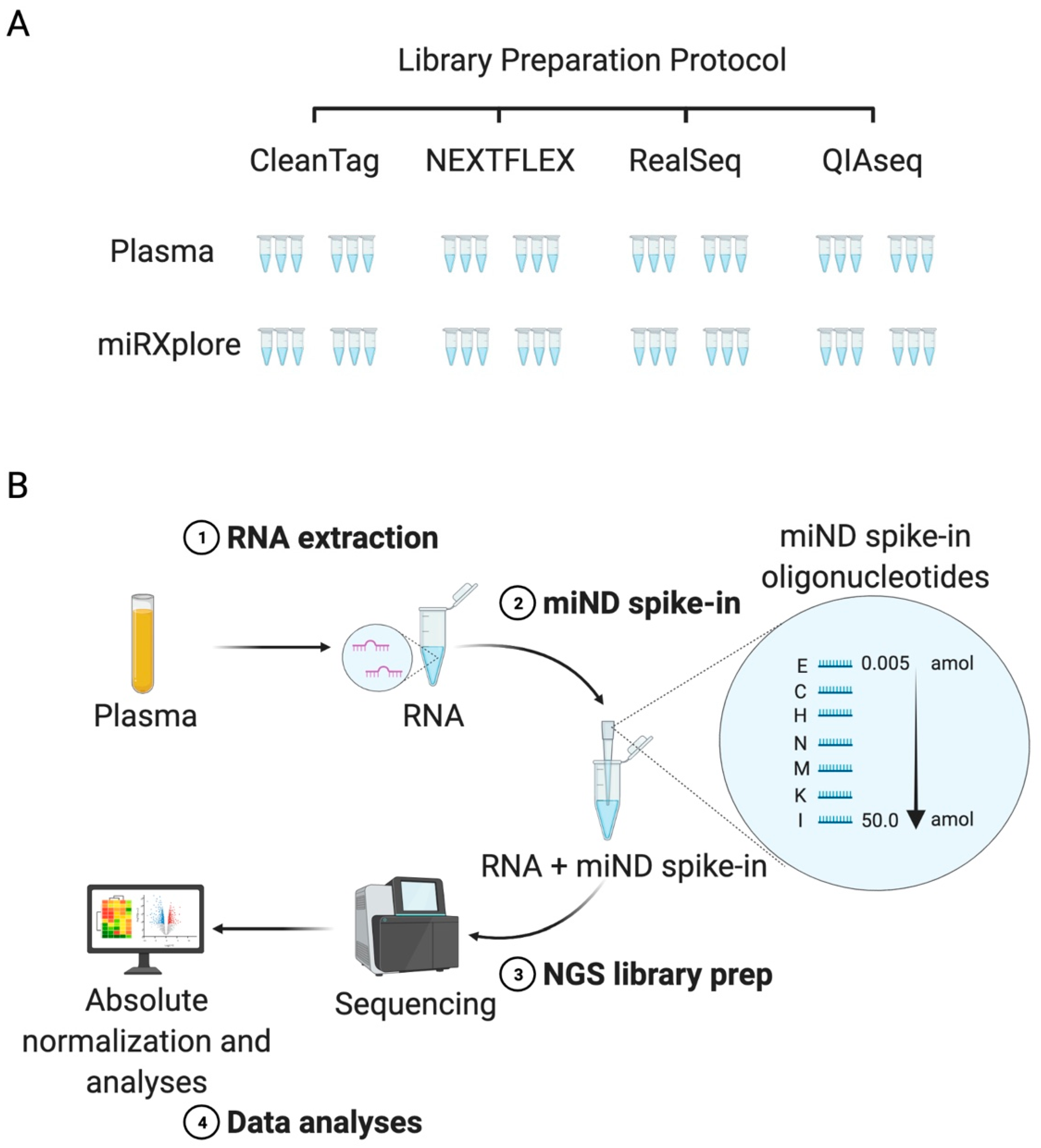
IJMS | Free Full-Text | A MicroRNA Next-Generation-Sequencing Discovery Assay (miND) for Genome-Scale Analysis and Absolute Quantitation of Circulating MicroRNA Biomarkers
Decoding of Superimposed Traces Produced by Direct Sequencing of Heterozygous Indels | PLOS Computational Biology