Schematic explanation of one-hot encoding, zero-padding and truncation... | Download Scientific Diagram
Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks

python - How to transform amino acid raw data to 3d tensor by one hot encoding by tensorflow - Stack Overflow

Neural networks to learn protein sequence–function relationships from deep mutational scanning data | PNAS
Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning
Genes | Free Full-Text | DeepNup: Prediction of Nucleosome Positioning from DNA Sequences Using Deep Neural Network
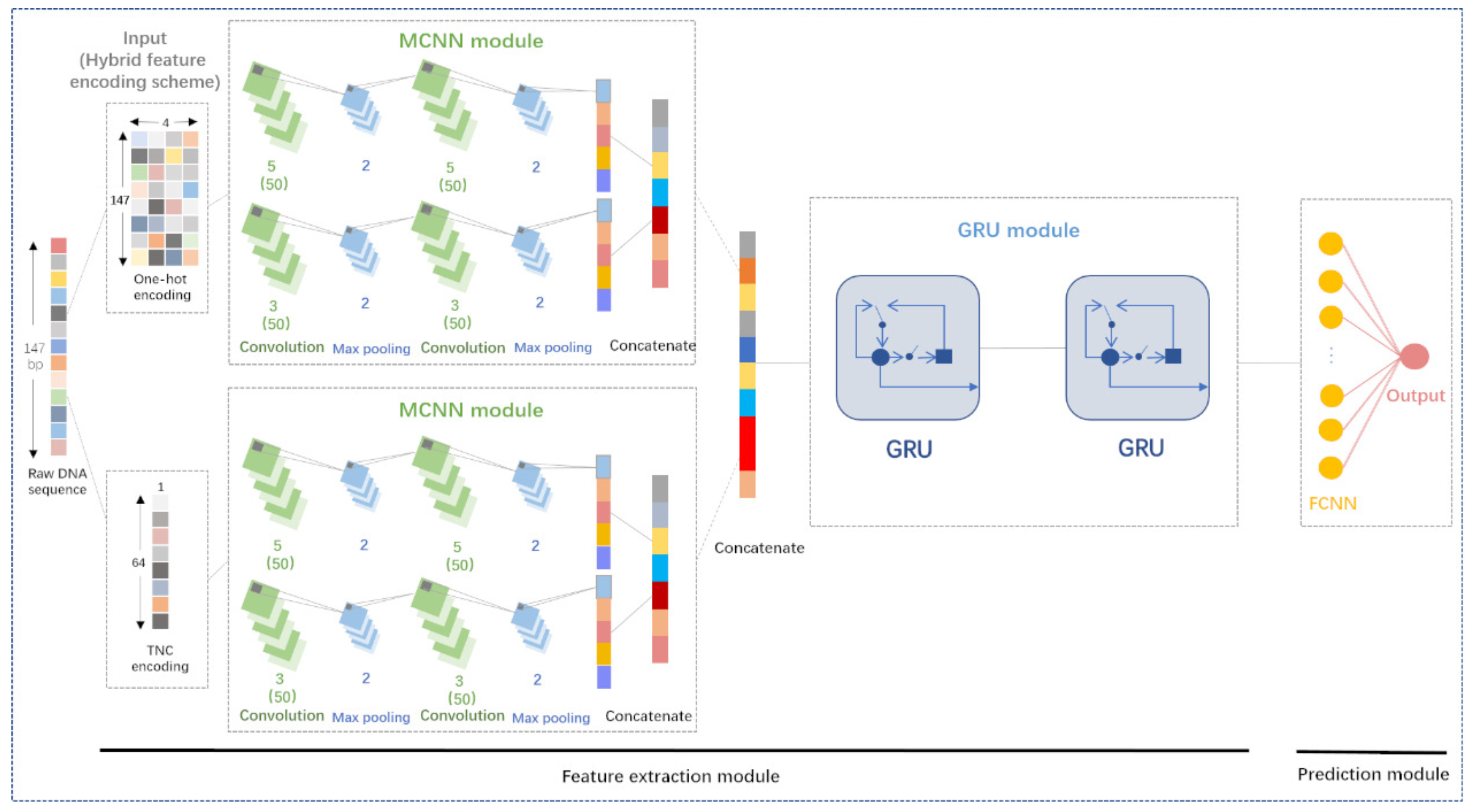
![Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ] Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/11262/1/fig-4-full.png)
Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ]
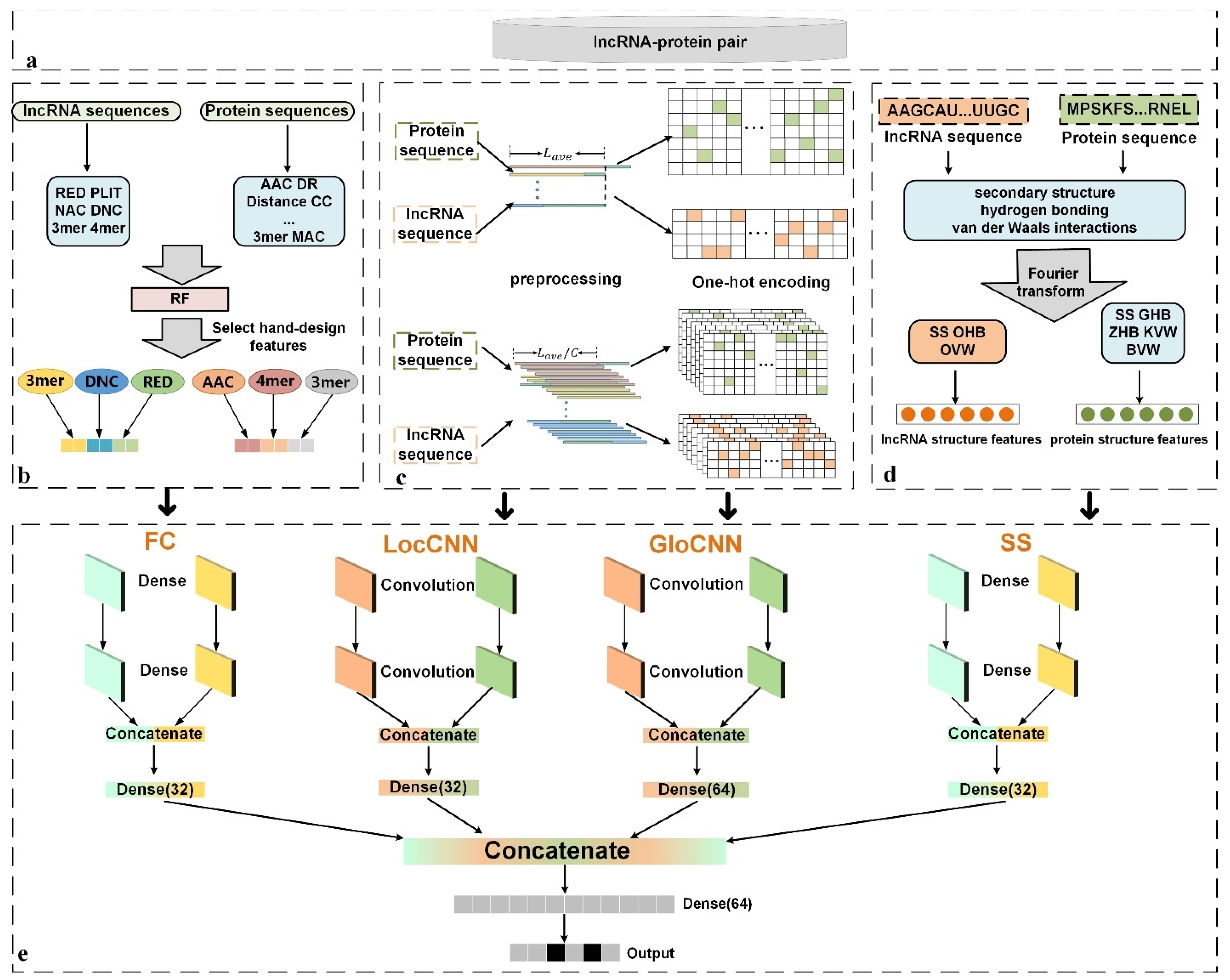
Genes | Free Full-Text | LGFC-CNN: Prediction of lncRNA-Protein Interactions by Using Multiple Types of Features through Deep Learning

Structure-aware protein solubility prediction from sequence through graph convolutional network and predicted contact map | Journal of Cheminformatics | Full Text

iRNA-PseKNC(2methyl): Identify RNA 2'-O-methylation sites by convolution neural network and Chou's pseudo components - ScienceDirect

DeepMHC: Deep Convolutional Neural Networks for High-performance peptide-MHC Binding Affinity Prediction | bioRxiv
Two common encoding methods. (A) One hot encoding of base, where black... | Download Scientific Diagram
![Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ] Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/11262/1/fig-1-2x.jpg)
Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ]
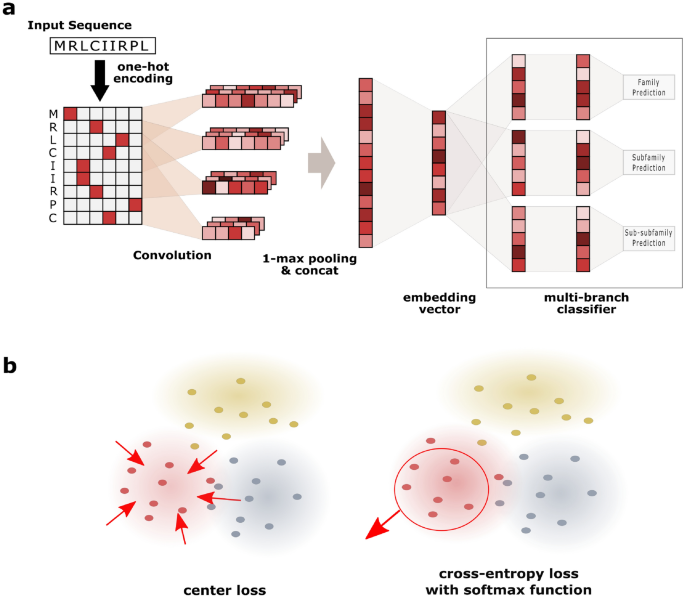
Deep hierarchical embedding for simultaneous modeling of GPCR proteins in a unified metric space | Scientific Reports

DNA / Protein Representation for Machine Learning Task with interactive code | by Jae Duk Seo | Towards Data Science
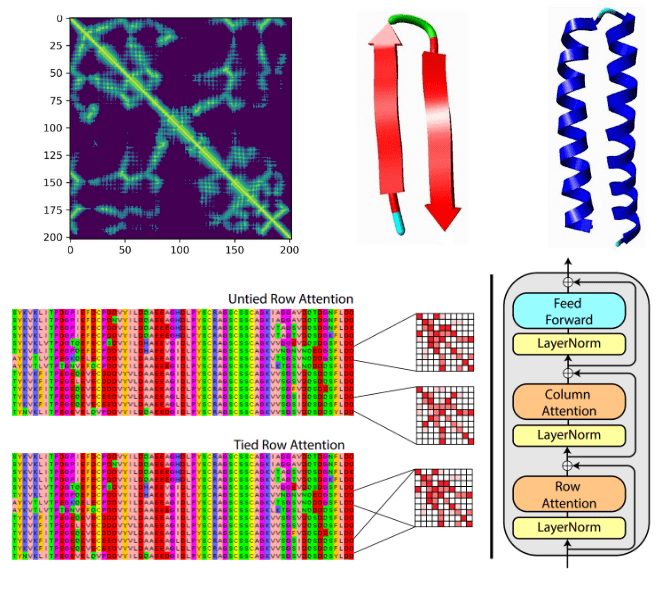
Deep learning on computational biology and bioinformatics tutorial: from DNA to protein folding and alphafold2 | AI Summer