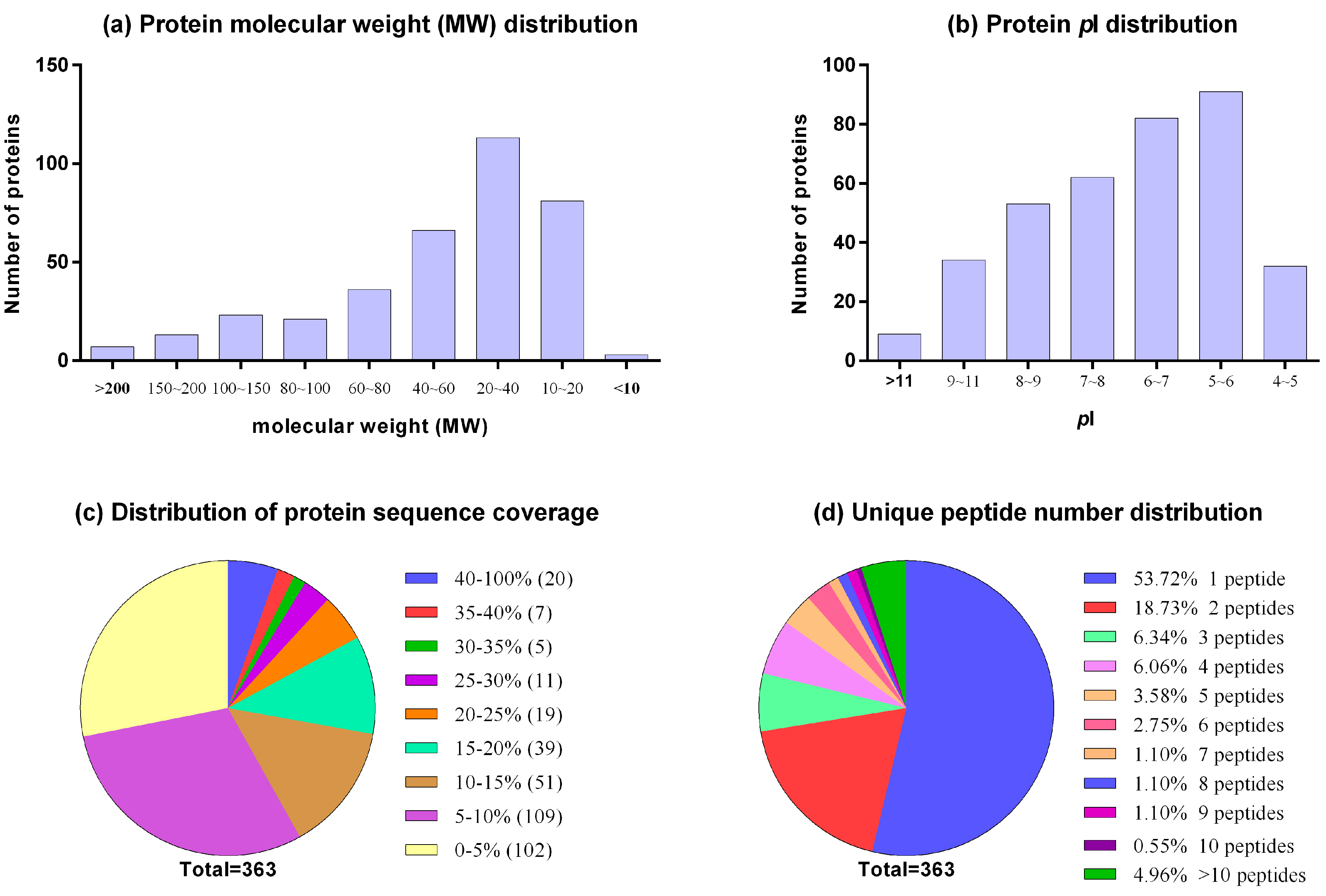
The distribution of protein's sequence coverage, number and length of... | Download Scientific Diagram

Maximizing Sequence Coverage in Top-Down Proteomics By Automated Multimodal Gas-Phase Protein Fragmentation | Analytical Chemistry

Figure 4. Sequence coverage of the observed protein spot svi002 peptides mapped to the known neoVTX β-subunit sequence. PMF matched peptides were shown with underlining. Further sequencing analysis was carried out by
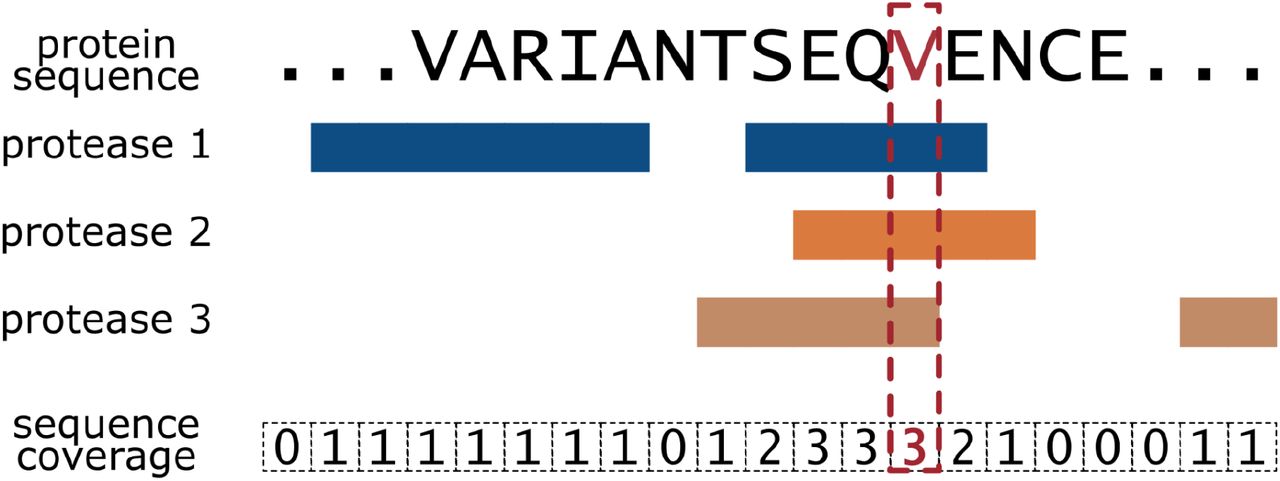
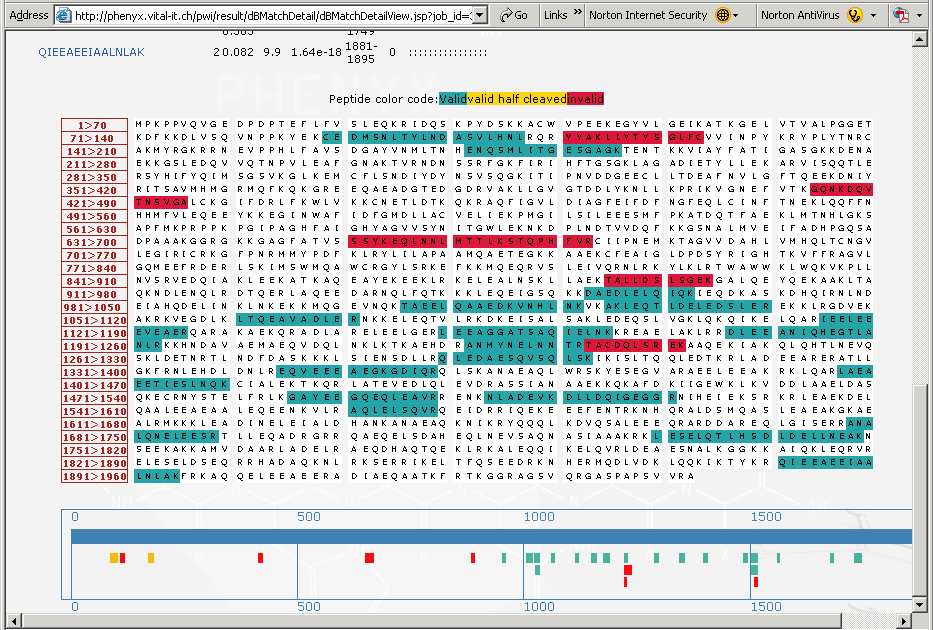
Validating amino acid variants in proteogenomics using sequence coverage by multiple reads | bioRxiv
Artificial fingerprints for cross-comparison of forensic DNA and protein recovery methods | PLOS ONE

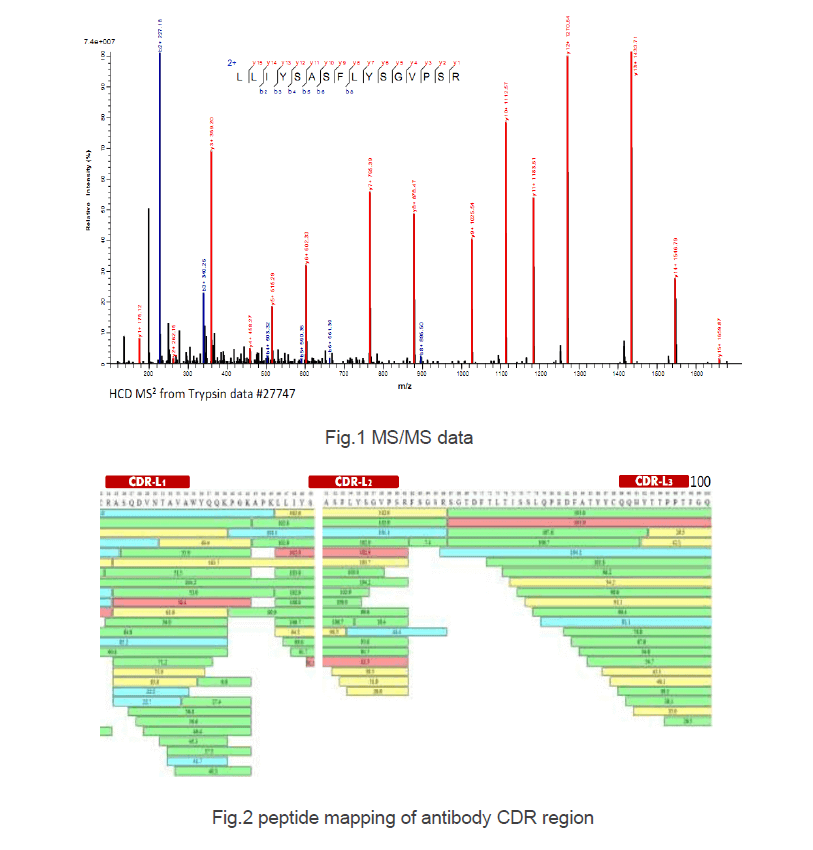
Internal Fragments Generated from Different Top-Down Mass Spectrometry Fragmentation Methods Extend Protein Sequence Coverage | Journal of the American Society for Mass Spectrometry

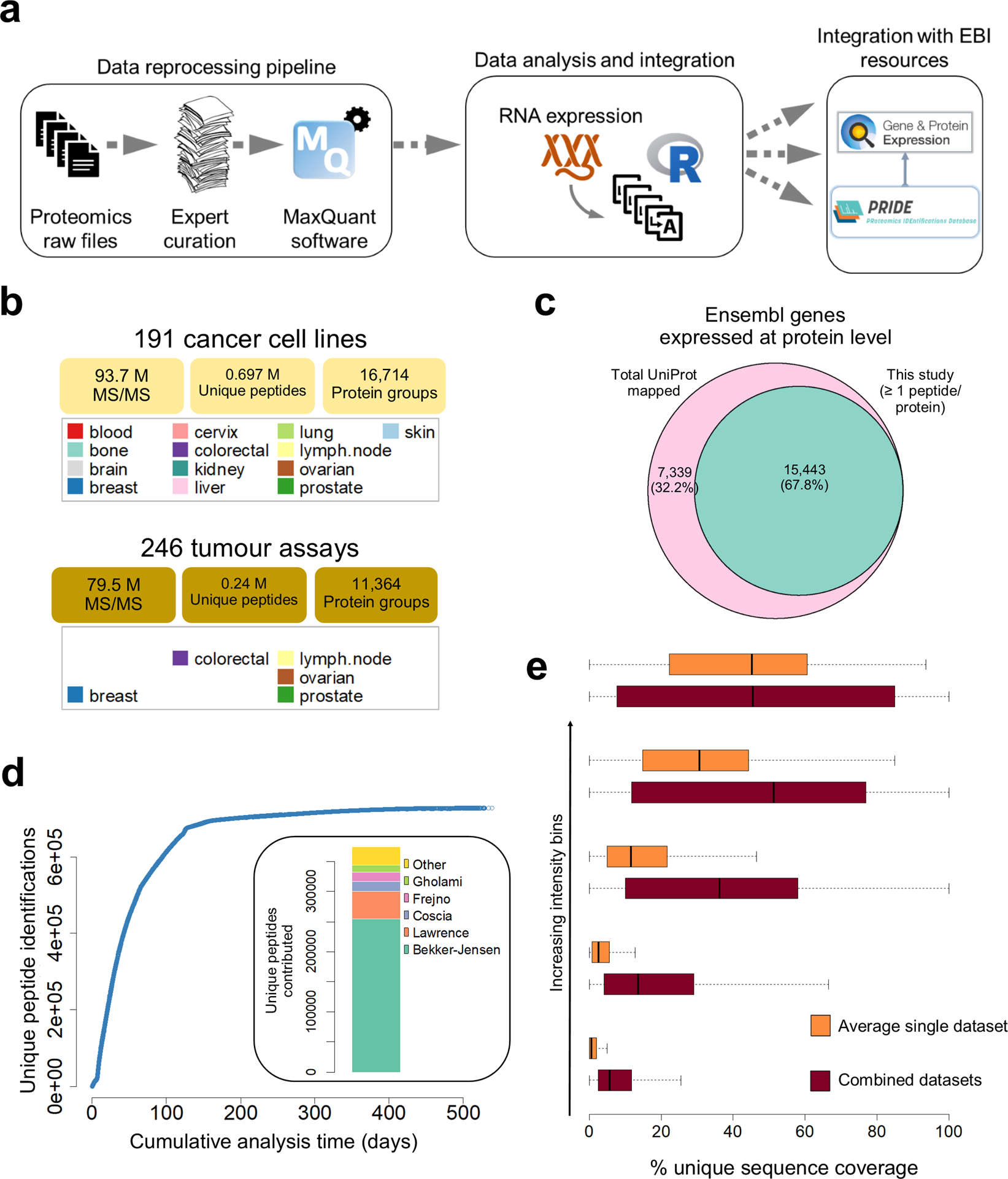
Data-independent acquisition protease-multiplexing enables increased proteome sequence coverage across multiple fragmentation modes | bioRxiv

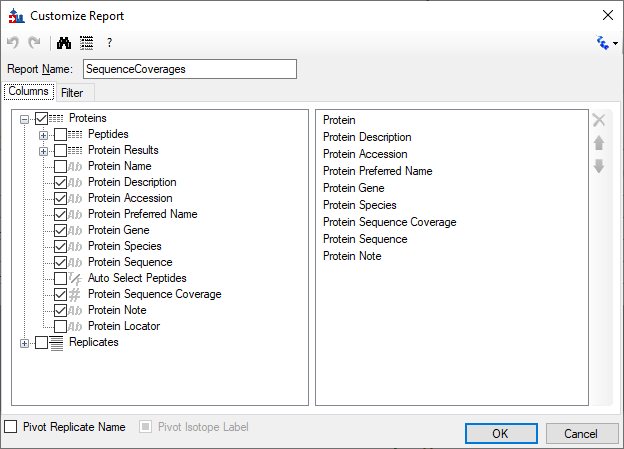
Sequence Coverage Visualizer: A Web Application for Protein Sequence Coverage 3D Visualization | Journal of Proteome Research
IJMS | Free Full-Text | Proteome Profile and Quantitative Proteomic Analysis of Buffalo (Bubalusbubalis) Follicular Fluid during Follicle Development

Proteome-sequence coverage for specimen Dm.5/157-16635 a, c, e, g, i,... | Download Scientific Diagram
![PDF] Less is More: Membrane Protein Digestion Beyond Urea–Trypsin Solution for Next-level Proteomics* | Semantic Scholar PDF] Less is More: Membrane Protein Digestion Beyond Urea–Trypsin Solution for Next-level Proteomics* | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a81bd8a6f3728fd88a166f95a81a58d268ffe283/10-Figure4-1.png)
PDF] Less is More: Membrane Protein Digestion Beyond Urea–Trypsin Solution for Next-level Proteomics* | Semantic Scholar

A method to identify and quantify the complete peptide composition in protein hydrolysates - ScienceDirect