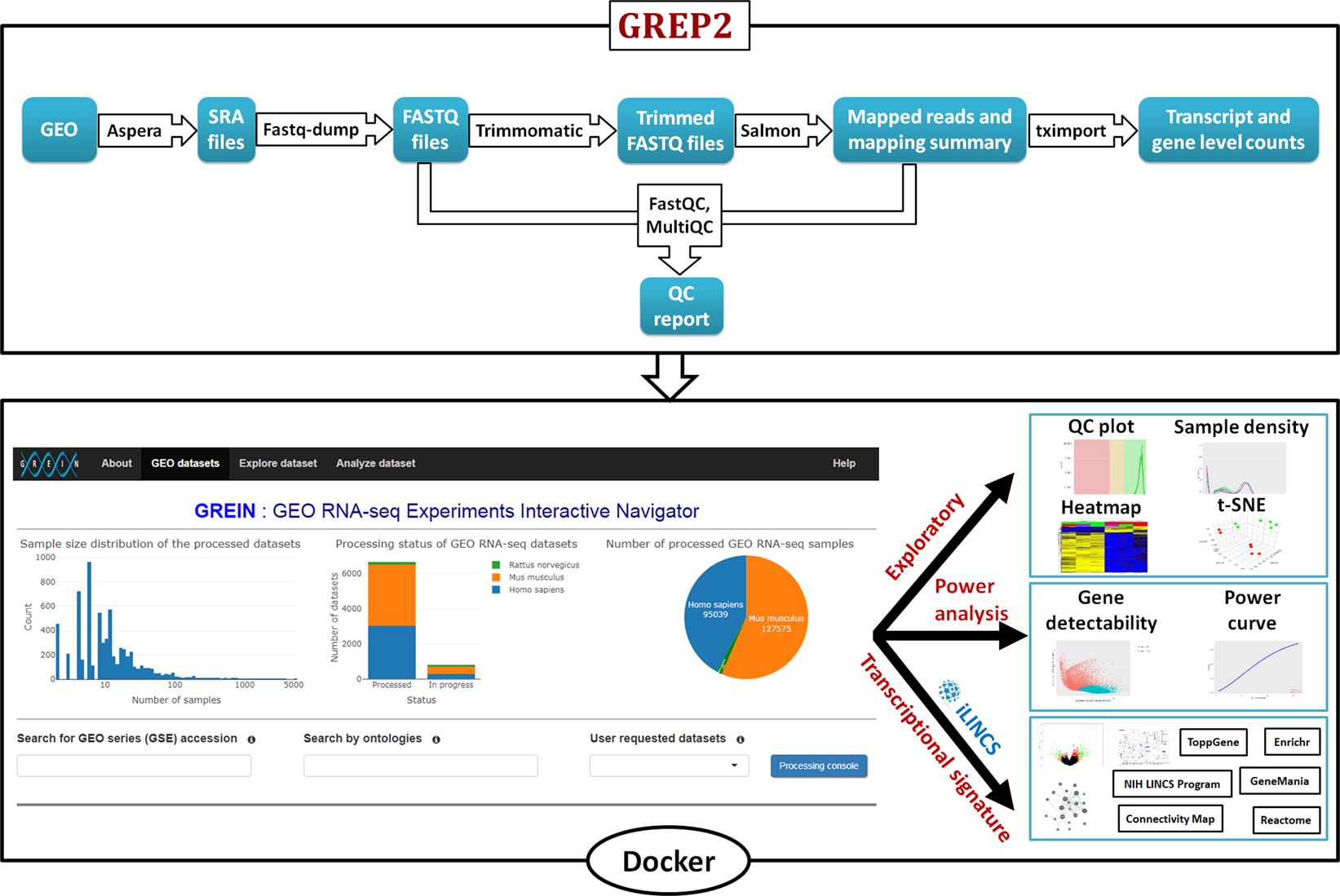
Dr. Aammar Tufail on Twitter: "Top 10 GitHub repositories for #RNAseq data analysis: A thread 🧵:" / Twitter
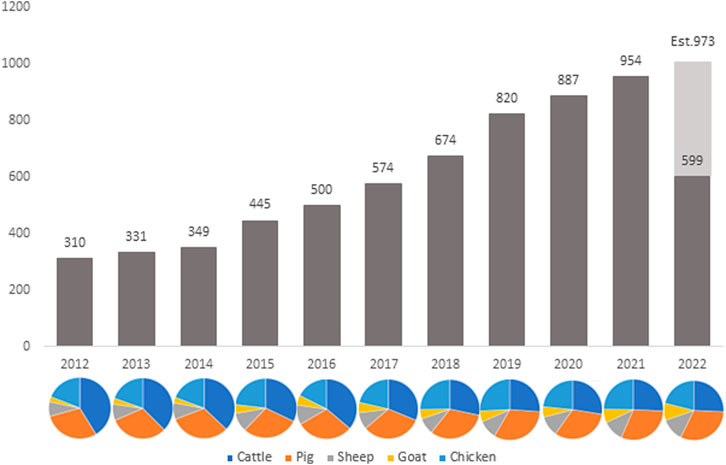
Frontiers | Recent developments and future directions in meta-analysis of differential gene expression in livestock RNA-Seq
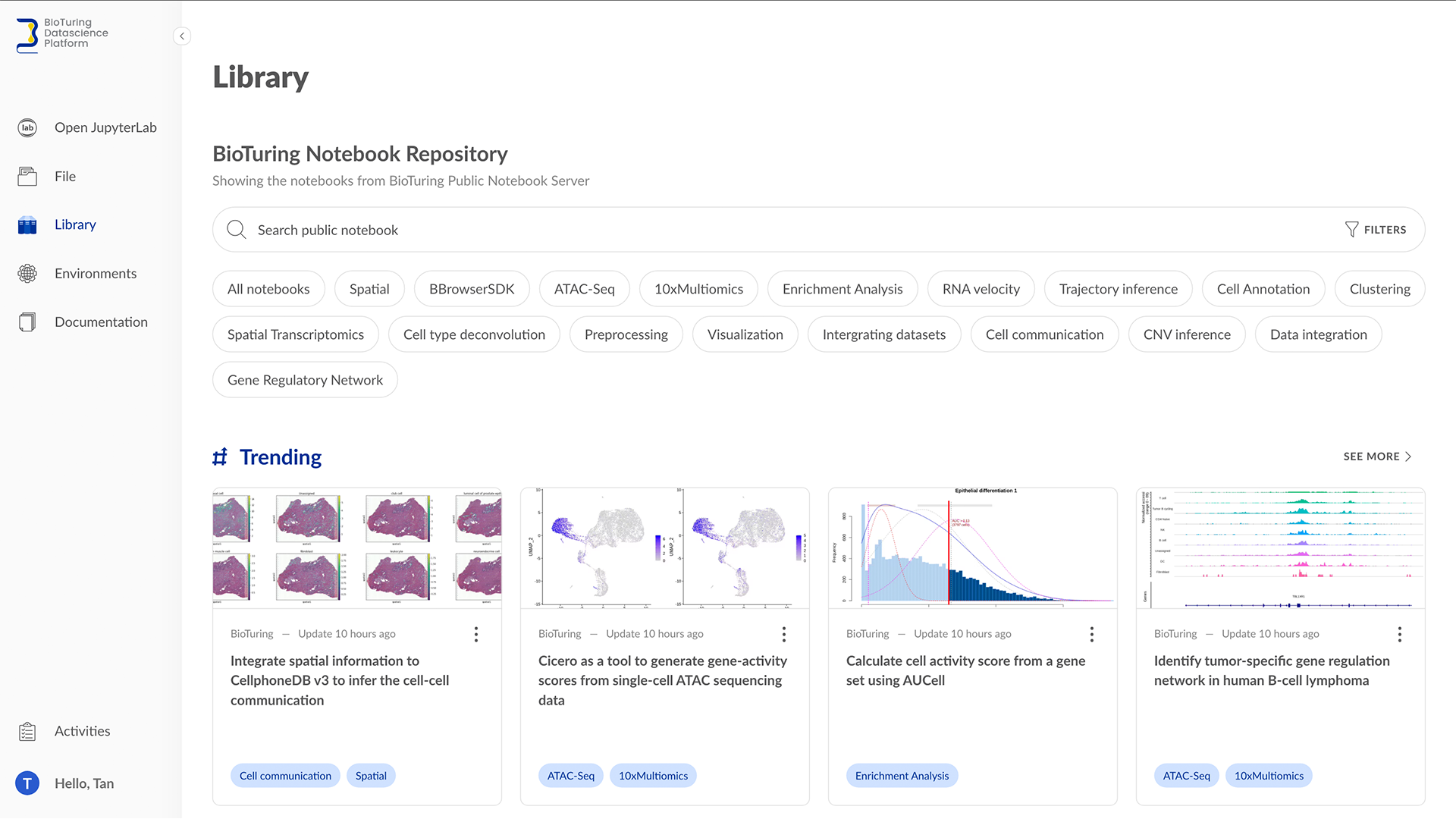
BioTuring Data Science Platform: A Jupyter notebook library of latest methods for single-cell analysis in R and Python - BioTuring's Blog

Quantitative and Qualitative RNA-Seq-Based Evaluation of Epstein-Barr Virus Transcription in Type I Latency Burkitt's Lymphoma Cells | Journal of Virology
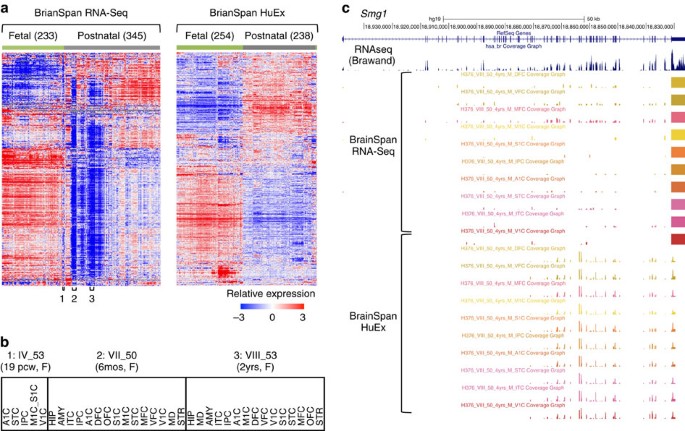
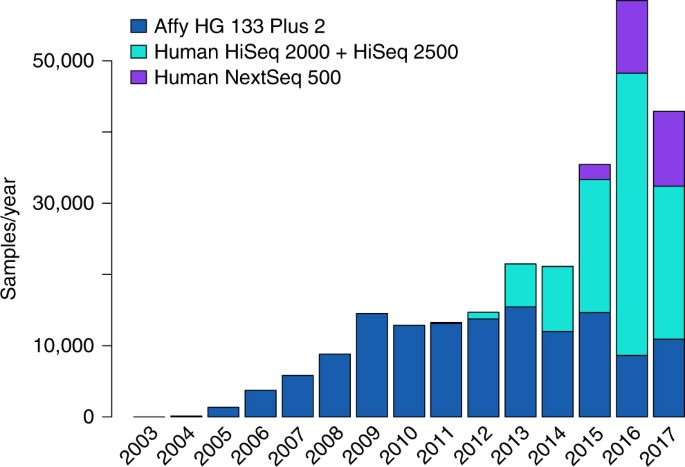
mRIN for direct assessment of genome-wide and gene-specific mRNA integrity from large-scale RNA-sequencing data | Nature Communications

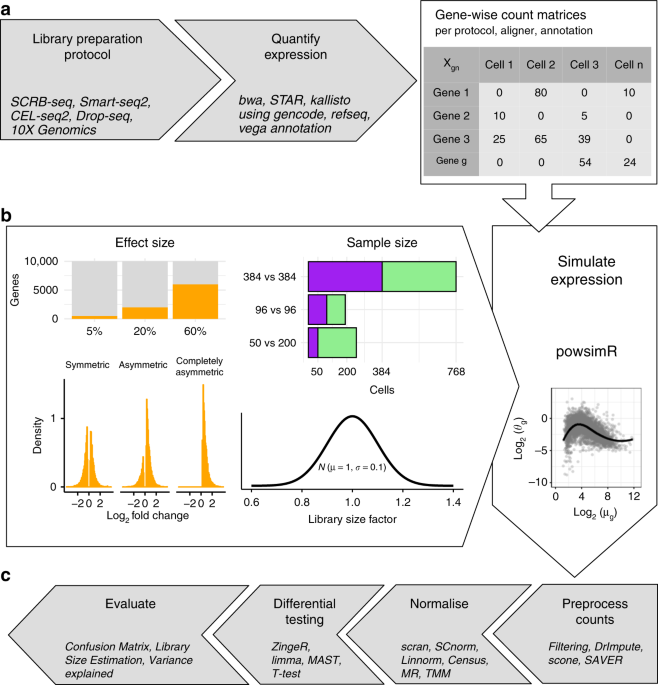
Cells | Free Full-Text | Detecting Interactive Gene Groups for Single-Cell RNA-Seq Data Based on Co-Expression Network Analysis and Subgraph Learning

A protocol to extract cell-type-specific signatures from differentially expressed genes in bulk-tissue RNA-seq - ScienceDirect

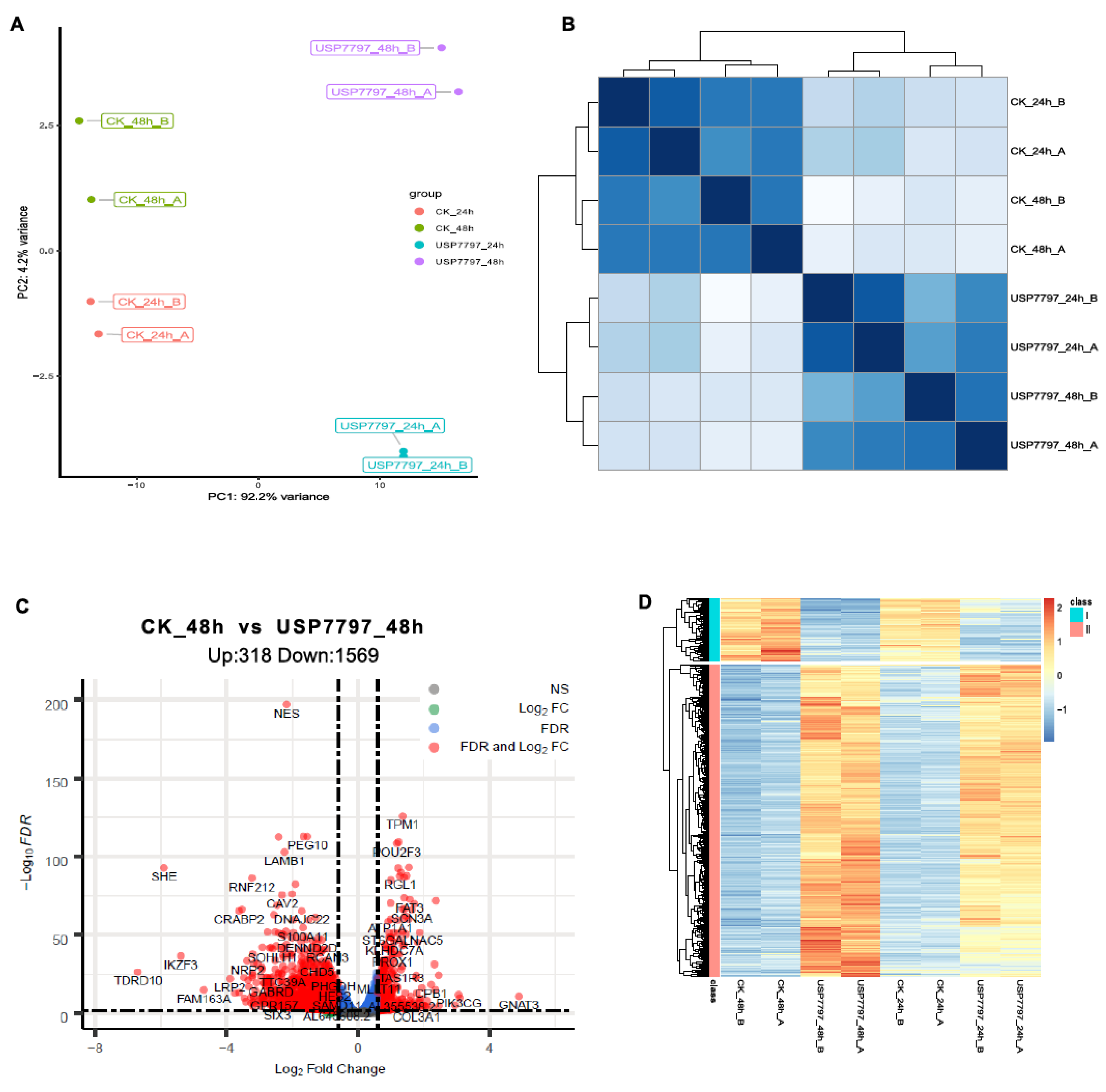
Transcriptomics based multi-dimensional characterization and drug screen in esophageal squamous cell carcinoma - eBioMedicine
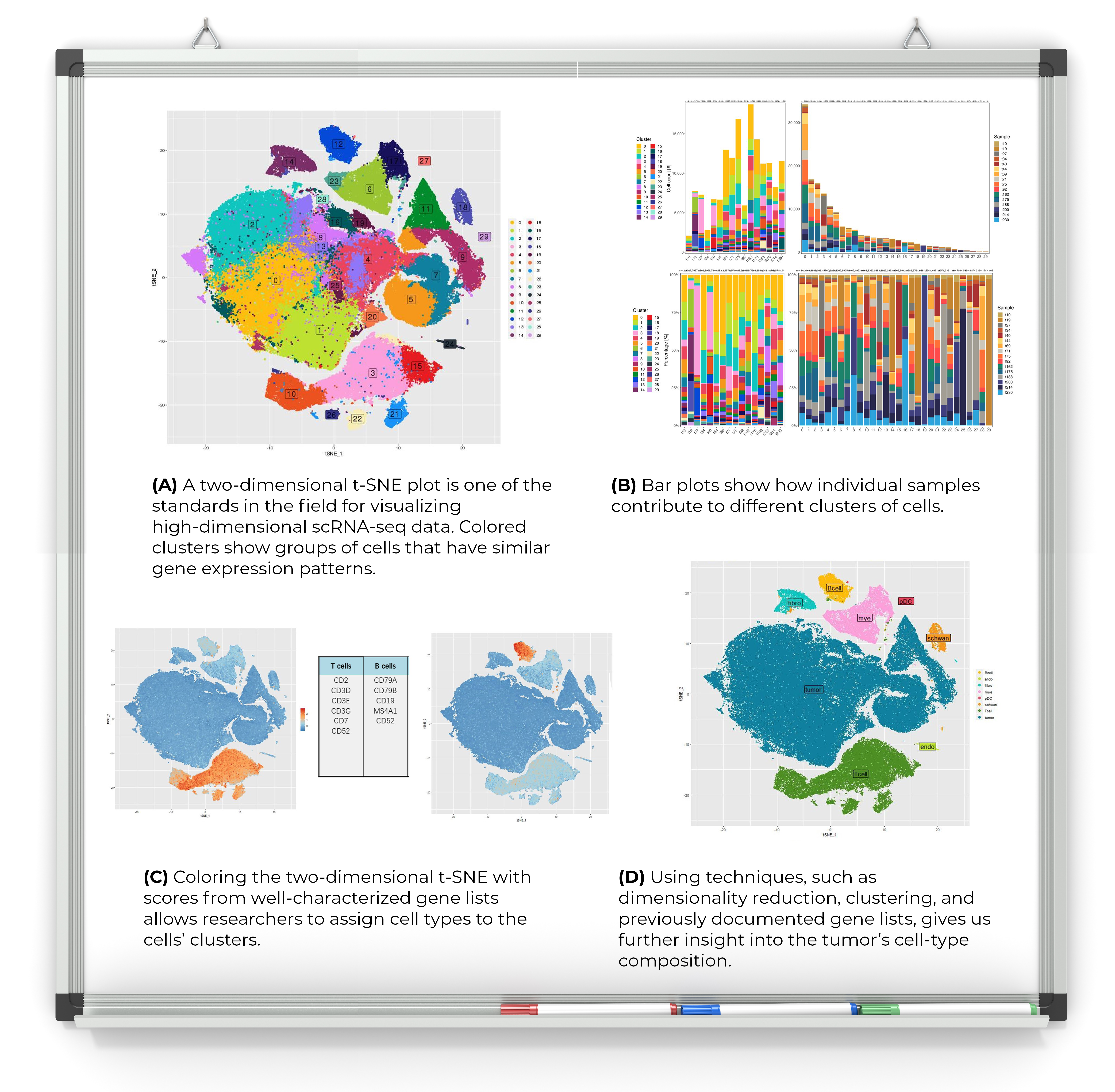
GitHub - theislab/single-cell-tutorial: Single cell current best practices tutorial case study for the paper:Luecken and Theis, "Current best practices in single-cell RNA-seq analysis: a tutorial"





![fRNAkenseq: a fully powered-by-CyVerse cloud integrated RNA-sequencing analysis tool [PeerJ] fRNAkenseq: a fully powered-by-CyVerse cloud integrated RNA-sequencing analysis tool [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2020/8592/1/fig-1-full.png)

![Detecting heterogeneity in single-cell RNA-Seq data by non-negative matrix factorization [PeerJ] Detecting heterogeneity in single-cell RNA-Seq data by non-negative matrix factorization [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2017/2888/1/fig-1-full.png)