Boxplot of sequencing depth data and amplicons size (bp). The range of... | Download Scientific Diagram
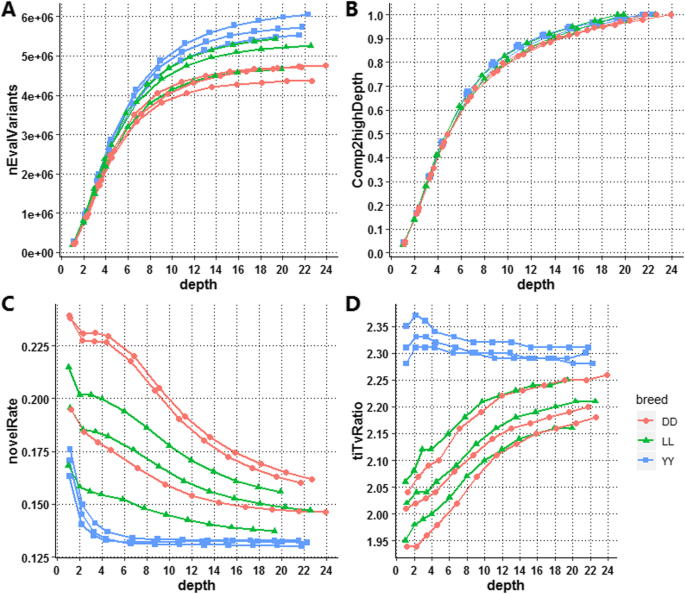
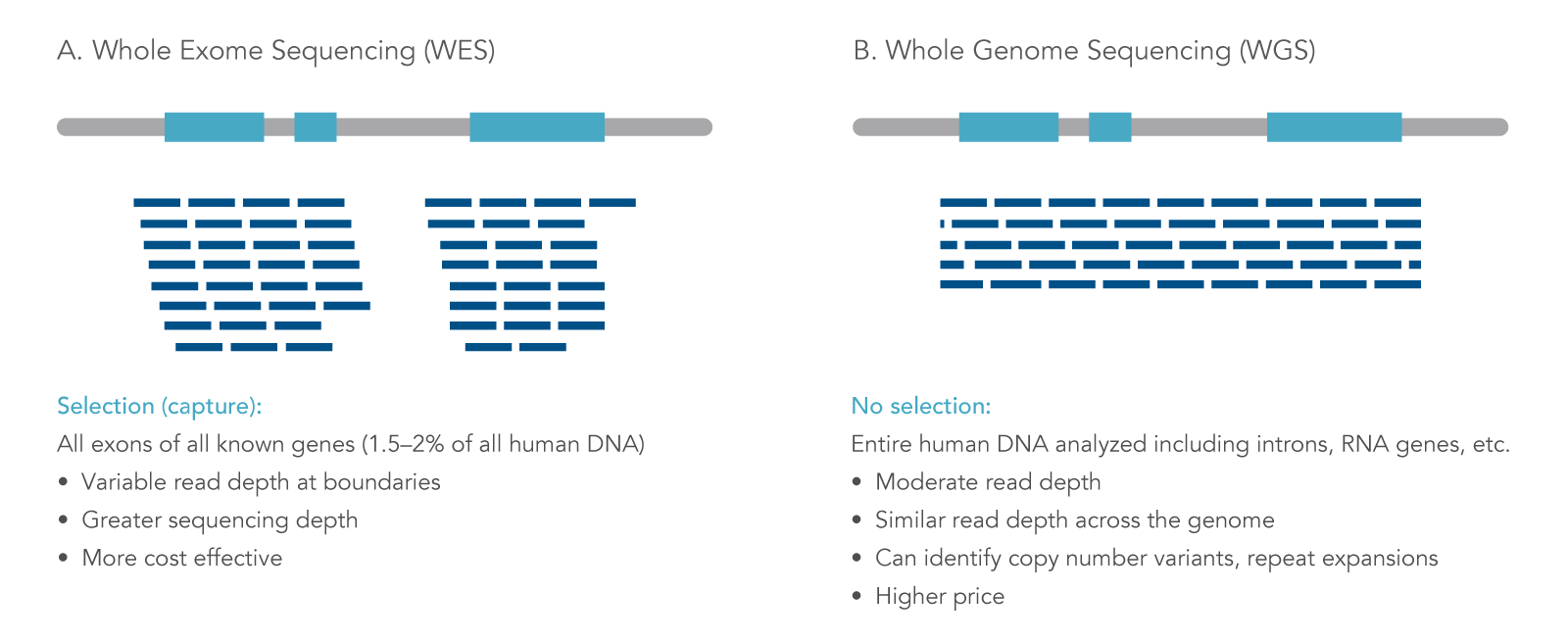
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text
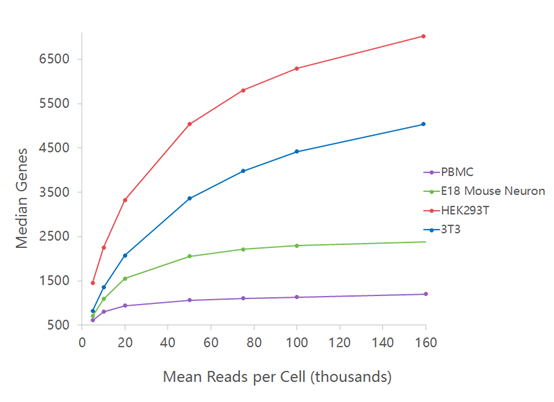
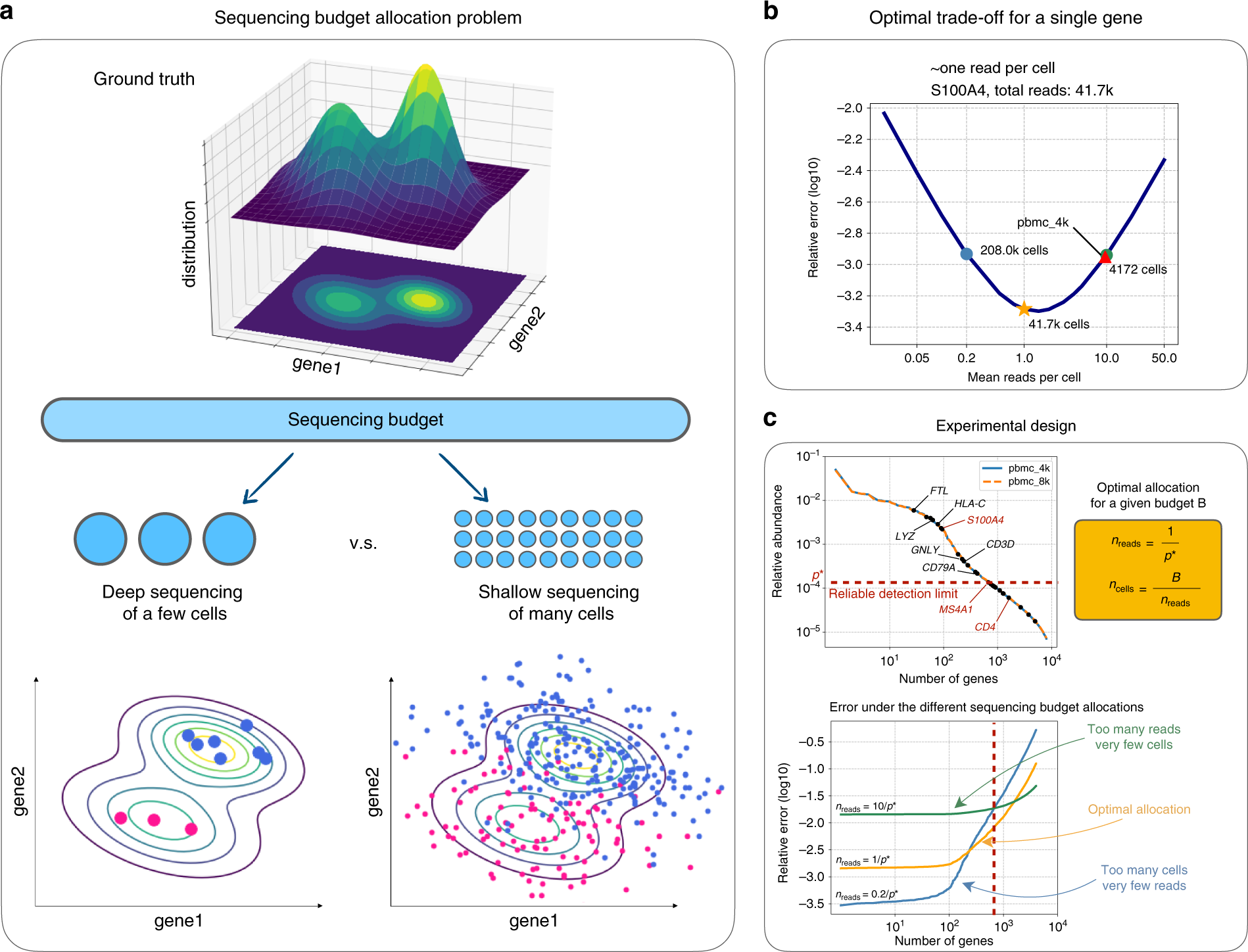
Single-cell RNA-seq: Normalization, identification of most variable genes | Introduction to single-cell RNA-seq
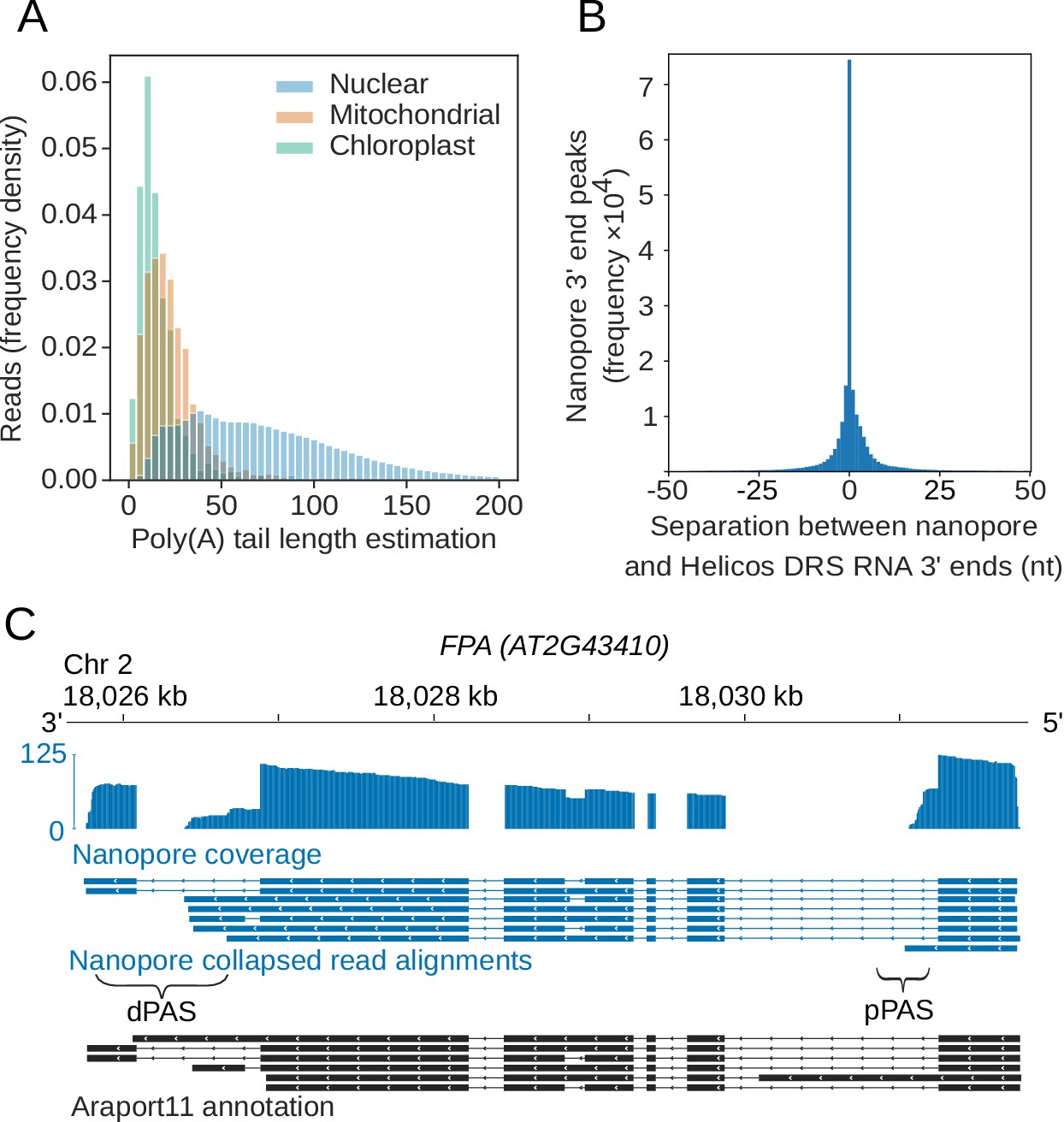
Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification | eLife

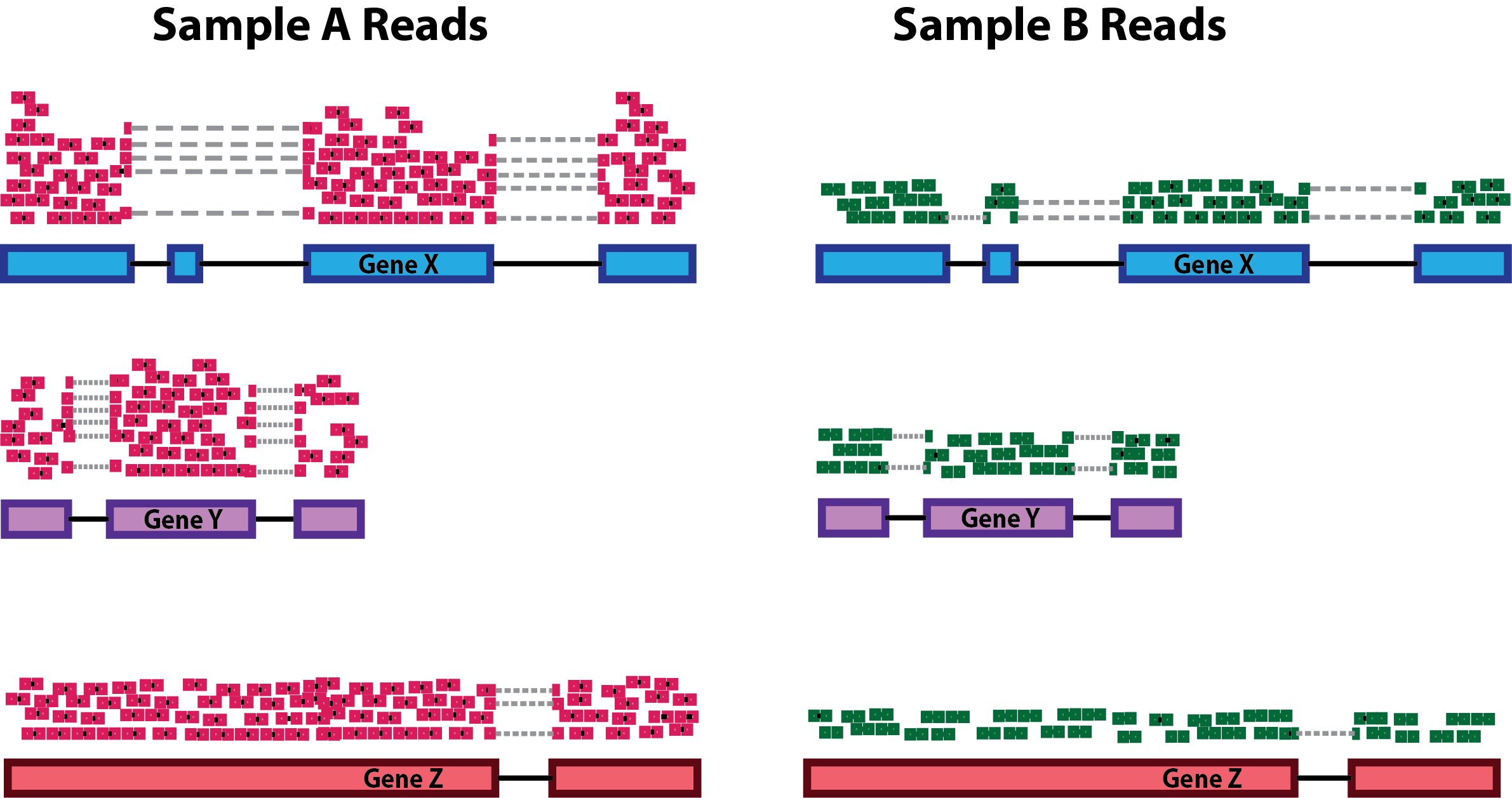
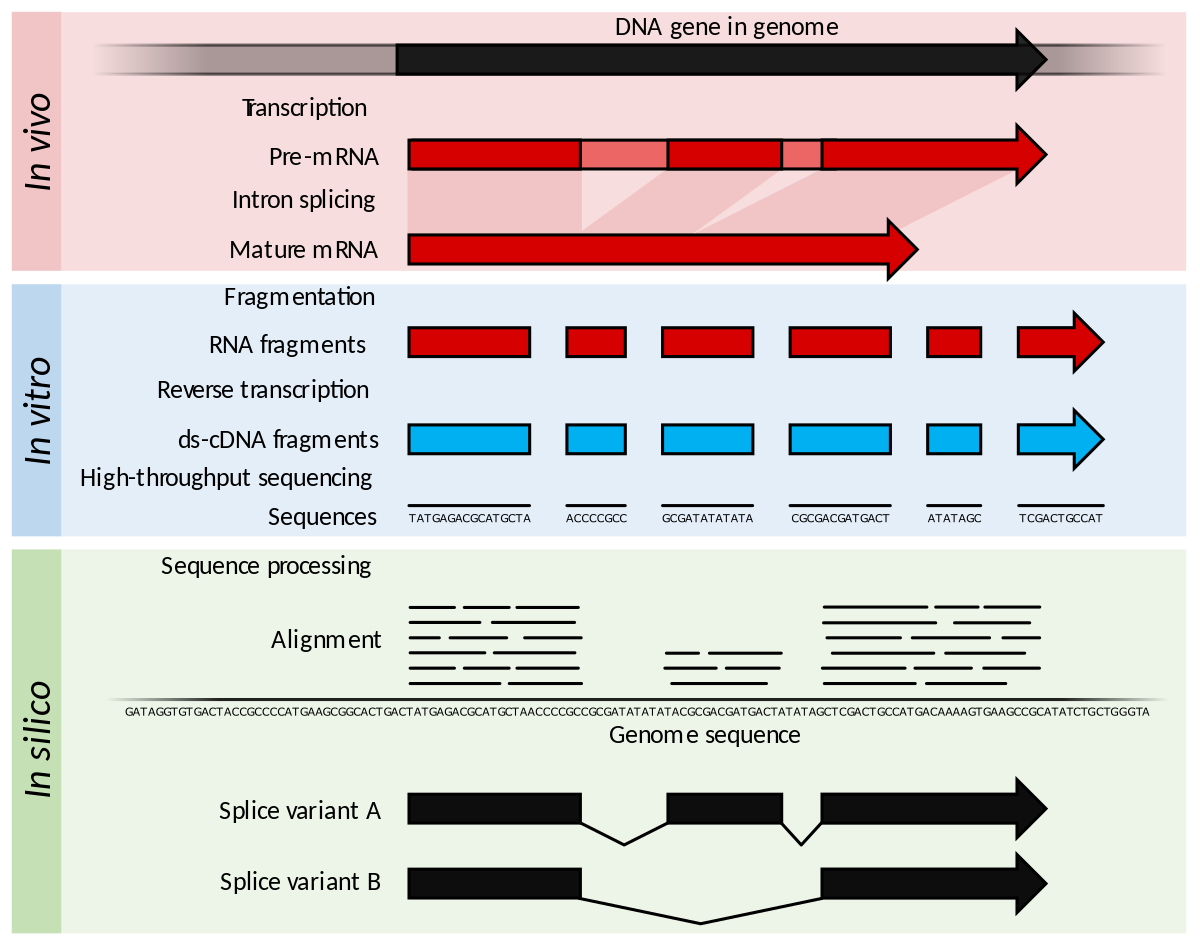
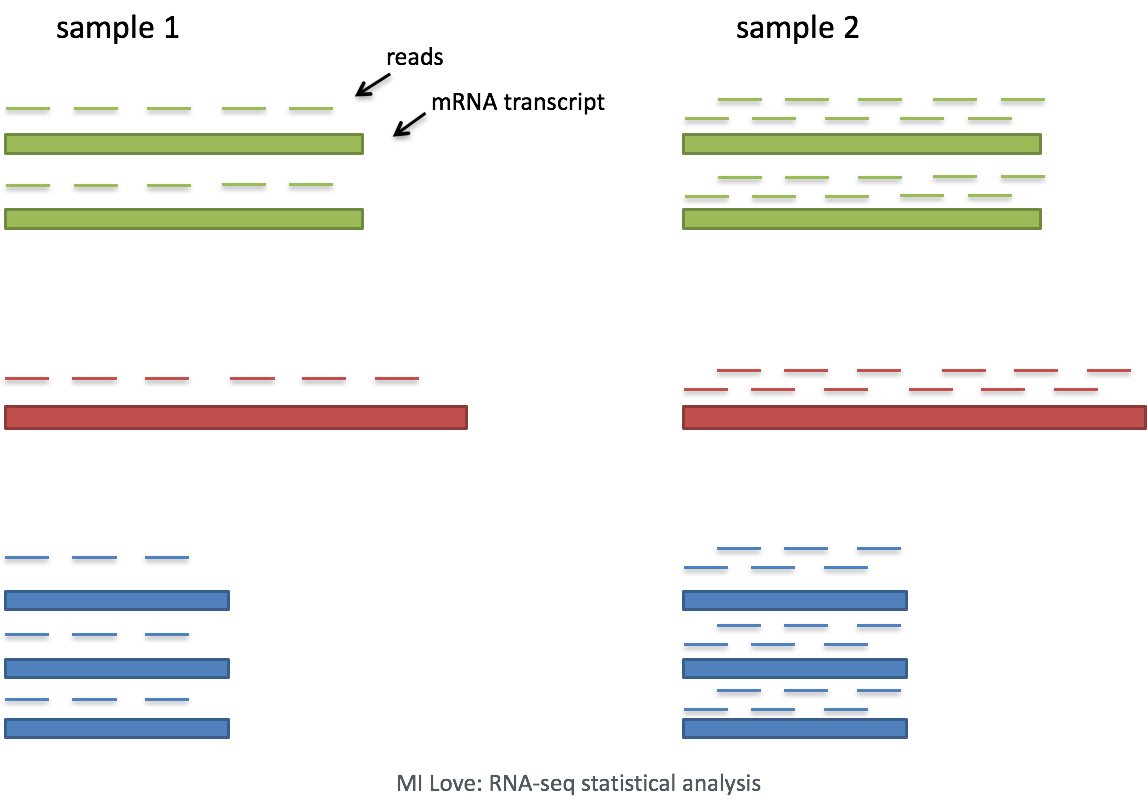
ScienceVision - Different RNA-Seq experiment types require different sequencing read lengths and depth (number of reads per sample). This bulletin reviews RNA sequencing considerations and offers resources for planning RNA-Seq experiments. Link:
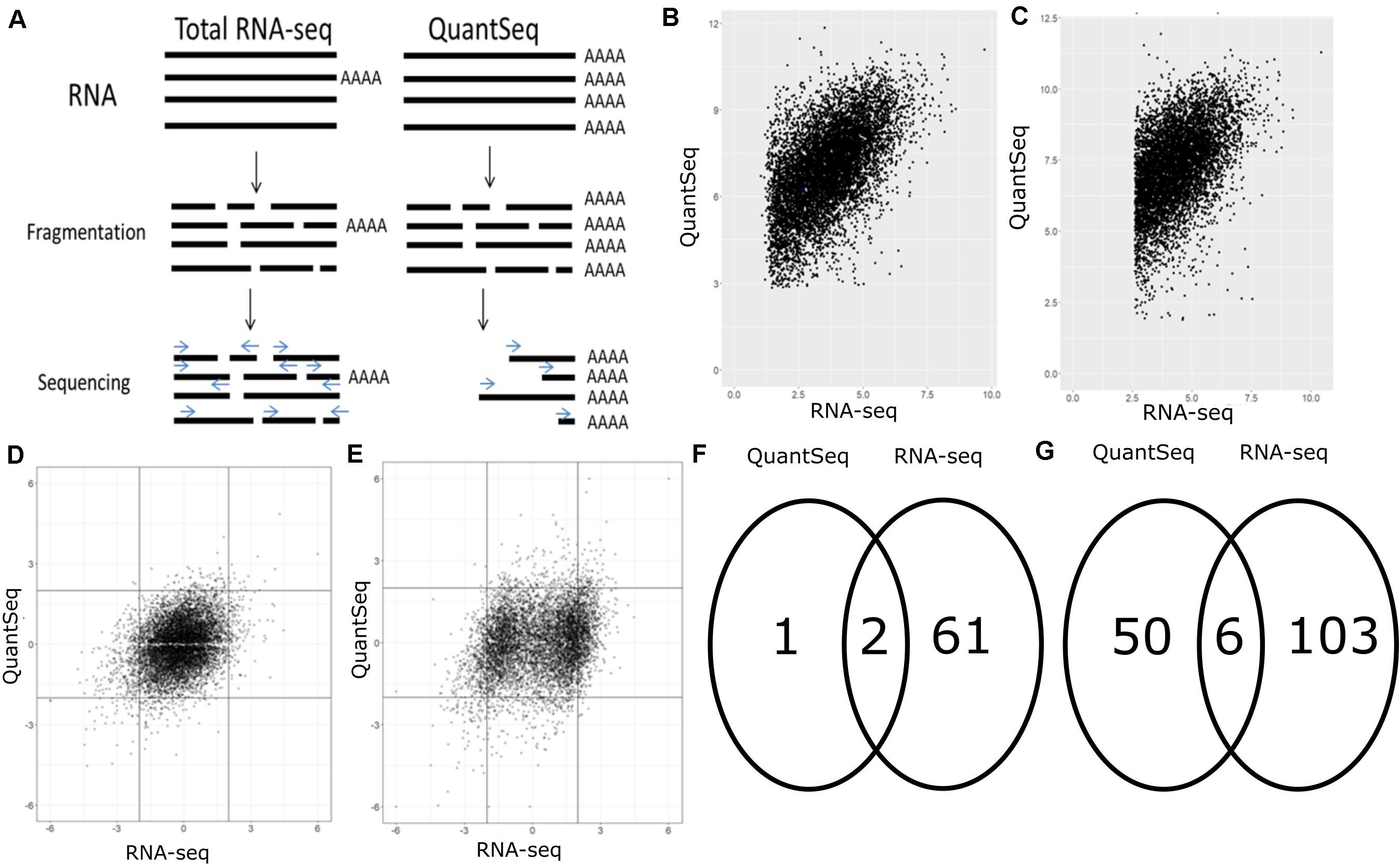
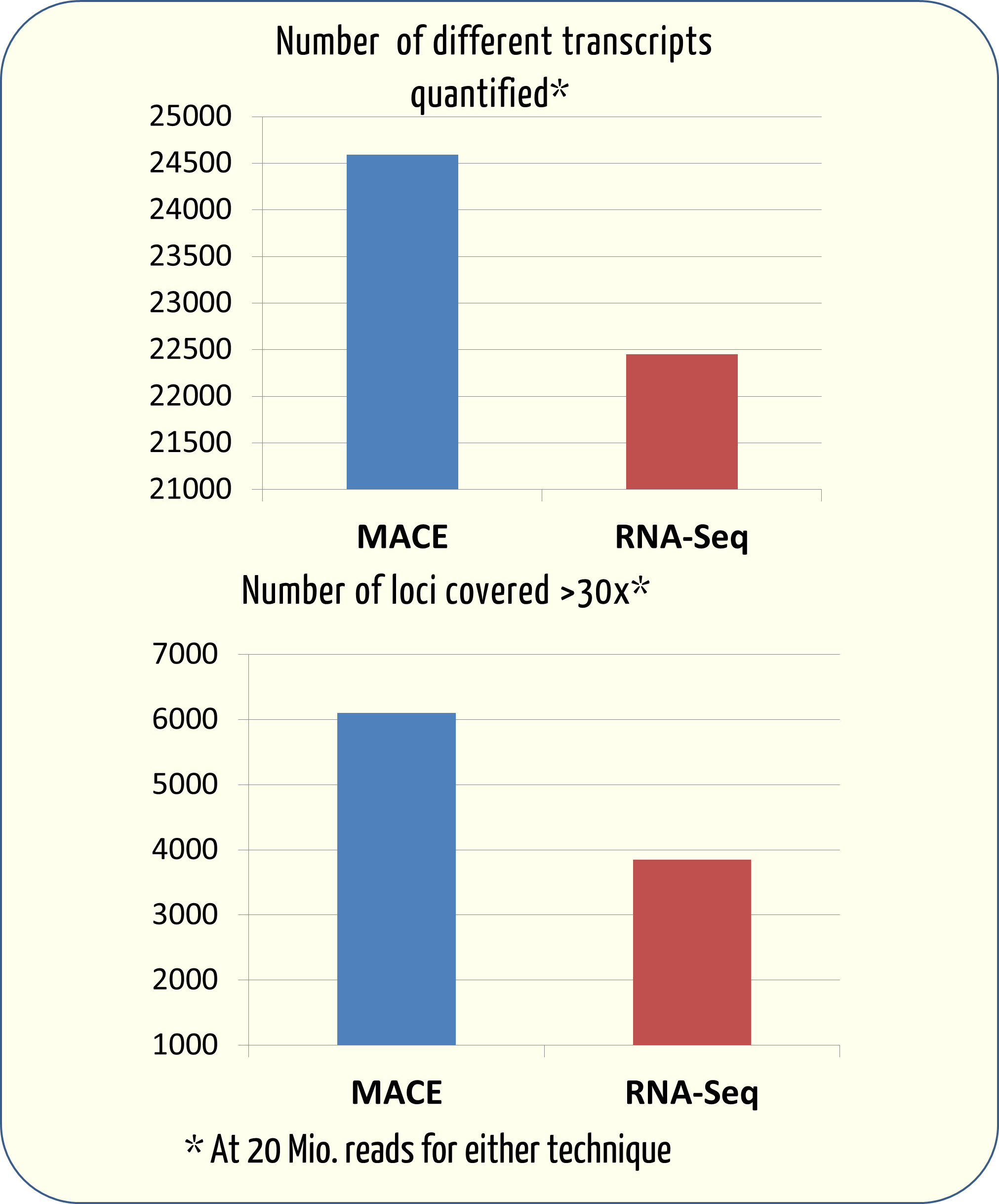
Frontiers | A Comparison of Low Read Depth QuantSeq 3′ Sequencing to Total RNA-Seq in FUS Mutant Mice