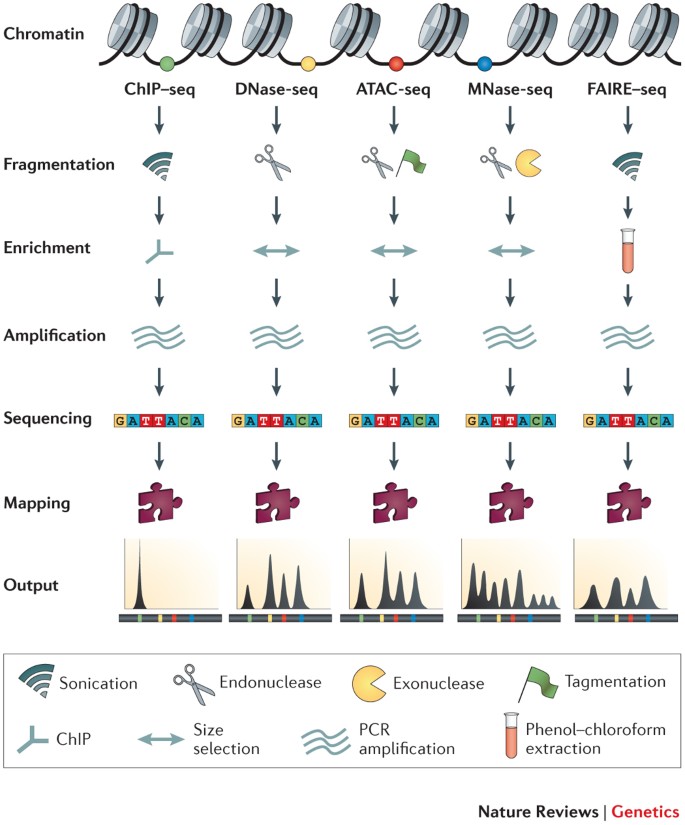
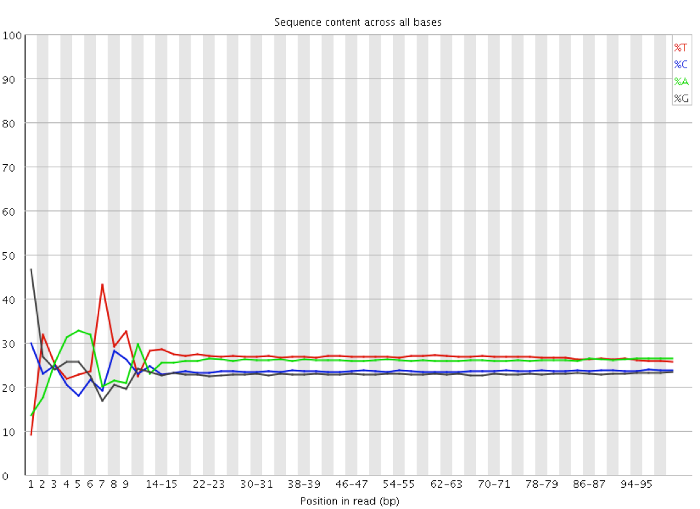
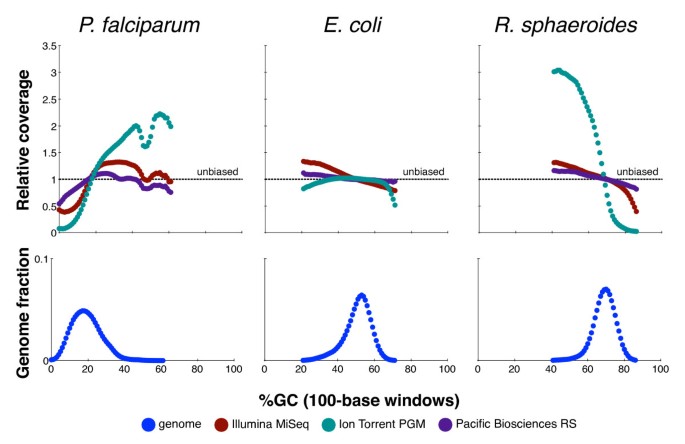
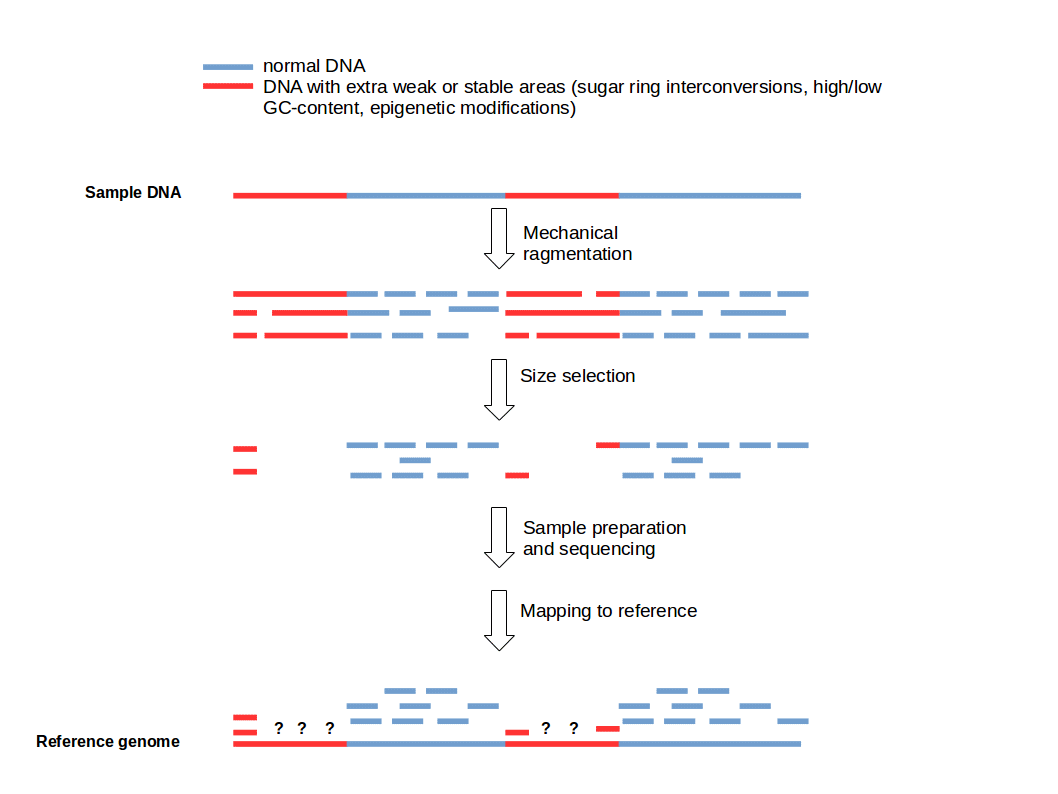Modeling of RNA-seq fragment sequence bias reduces systematic errors in transcript abundance estimation | Nature Biotechnology
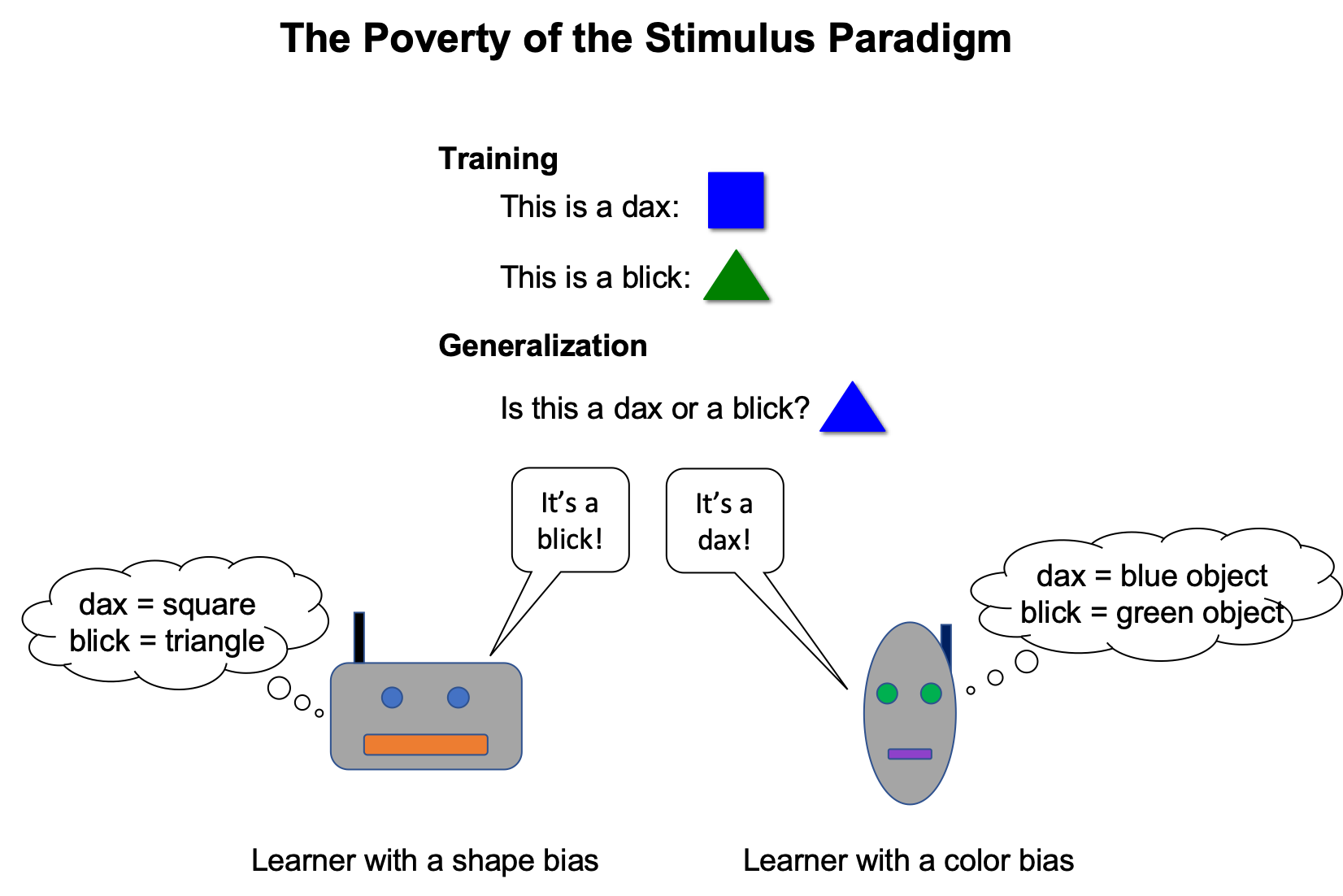
Does Syntax Need to Grow on Trees? Sources of Hierarchical Inductive Bias in Sequence-to-Sequence Networks
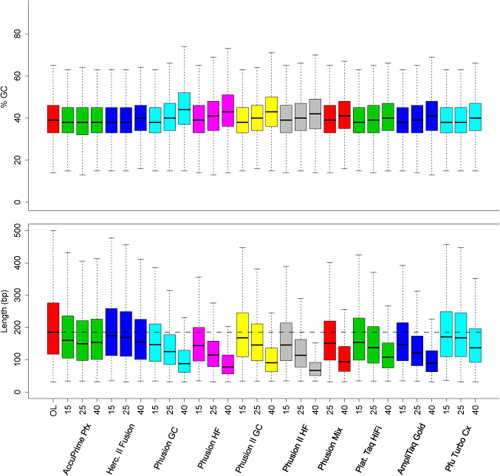
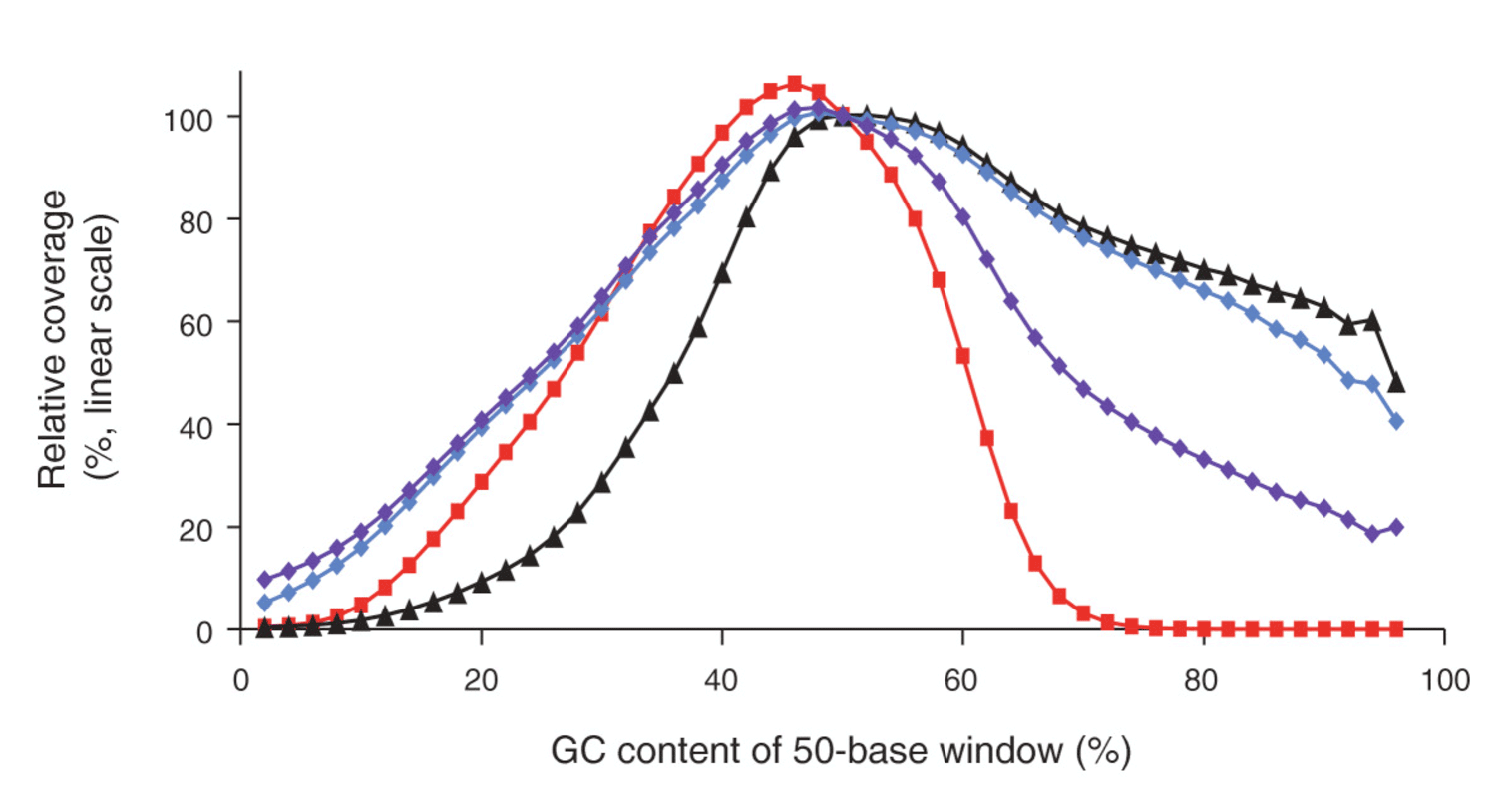
Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries | BioTechniques
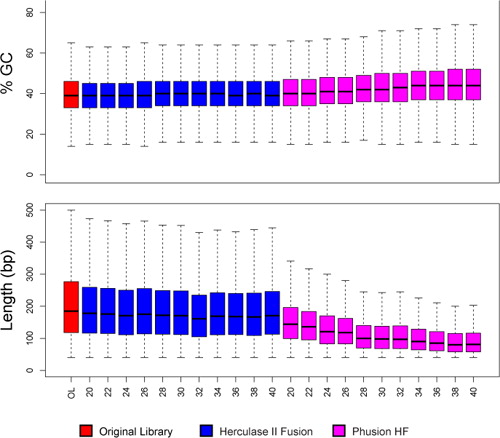
Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries | BioTechniques

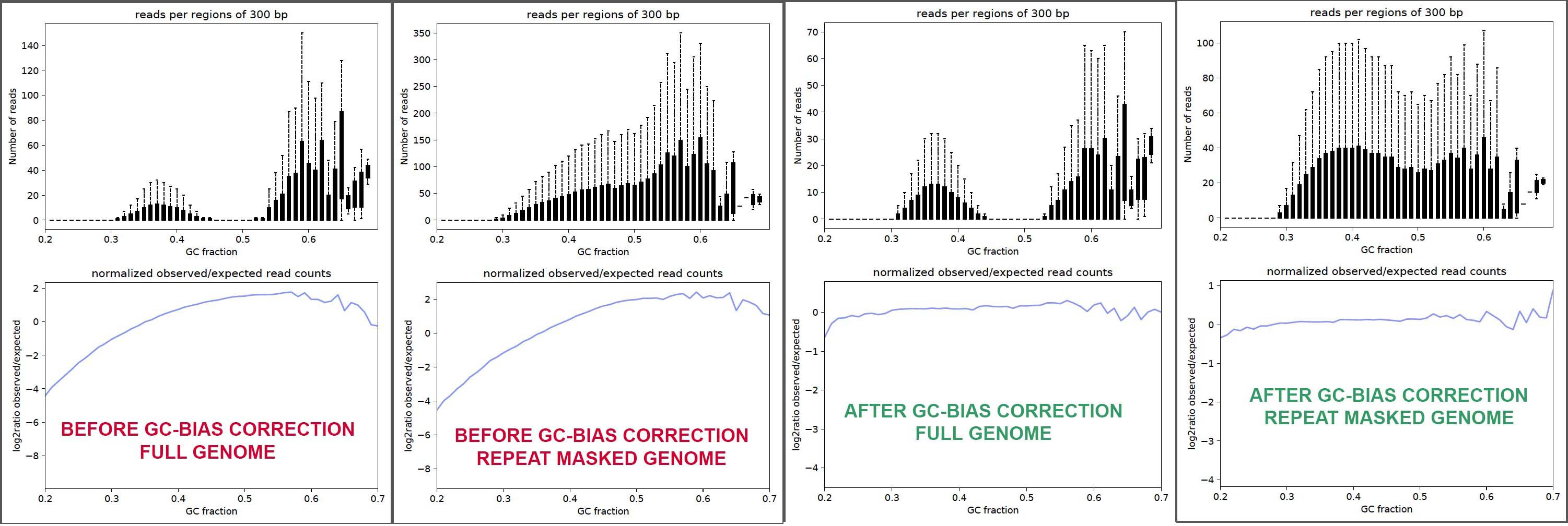
Sensitive and Low-Bias Transcriptome Sequencing Using Agarose PCR | ACS Applied Materials & Interfaces