Improved Annotation of 3′ Untranslated Regions and Complex Loci by Combination of Strand-Specific Direct RNA Sequencing, RNA-Seq and ESTs | PLOS ONE
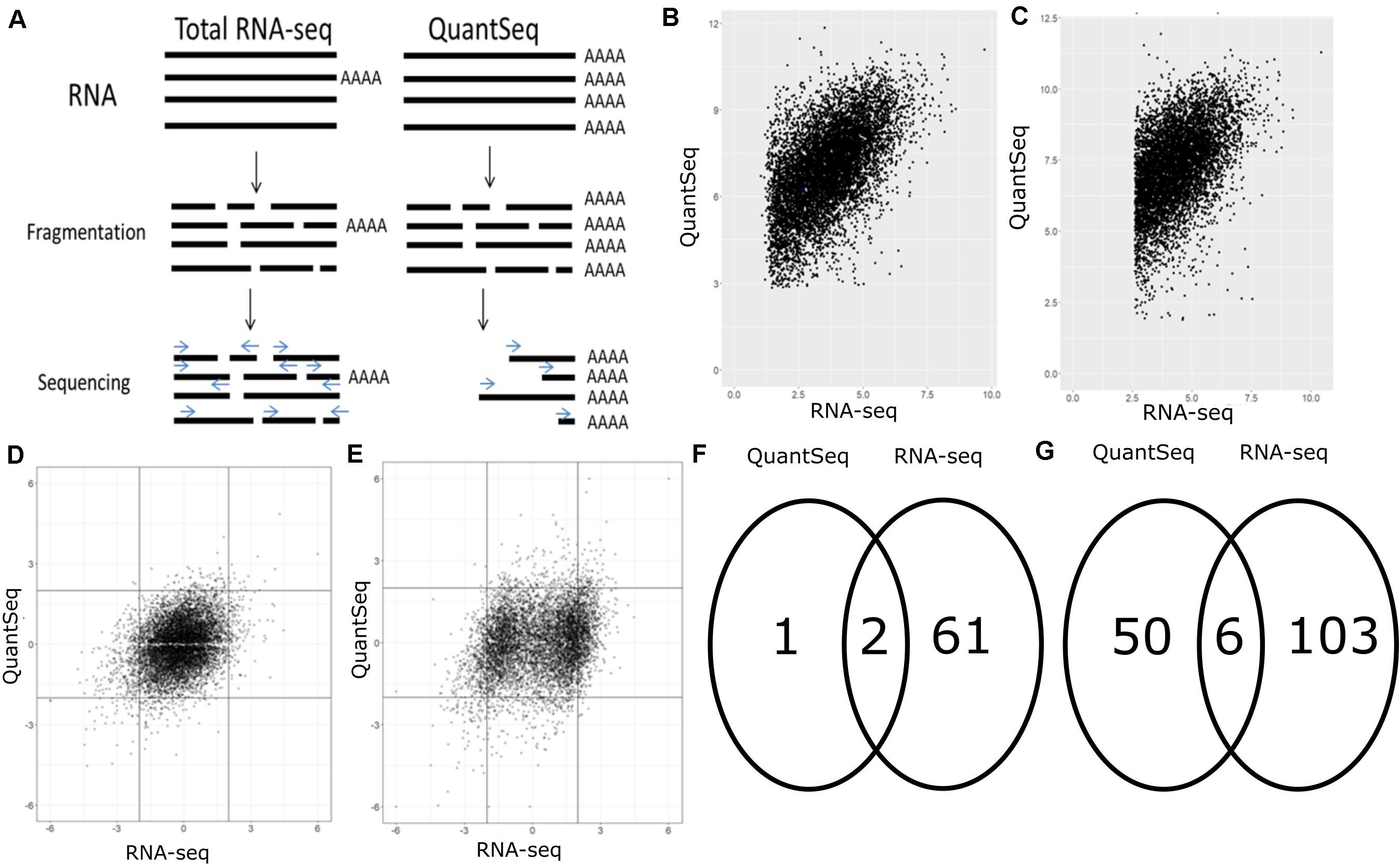
Frontiers | A Comparison of Low Read Depth QuantSeq 3′ Sequencing to Total RNA-Seq in FUS Mutant Mice
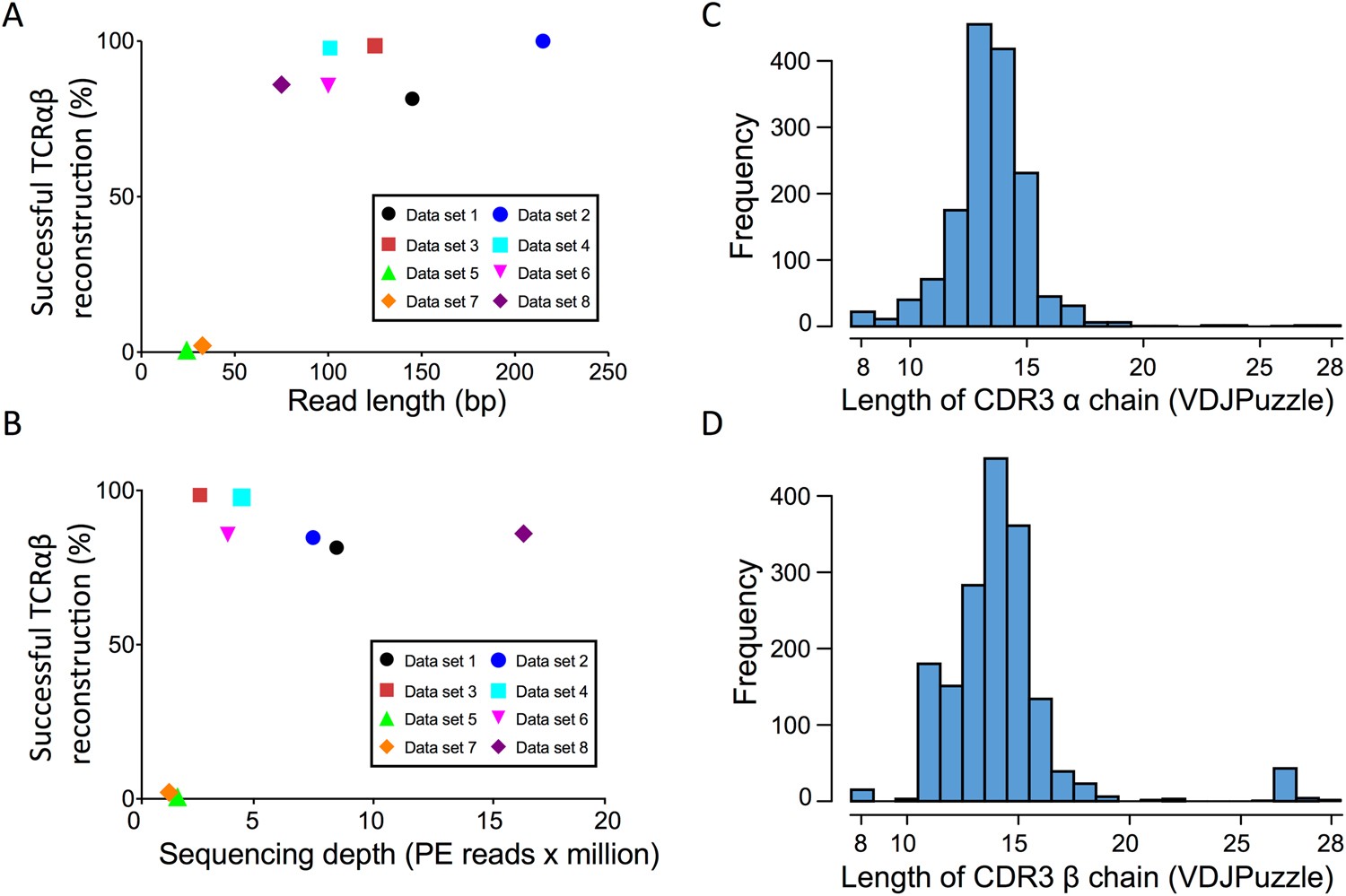
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

Determination of the number of reads needed for each RNA-Seq protocol... | Download Scientific Diagram