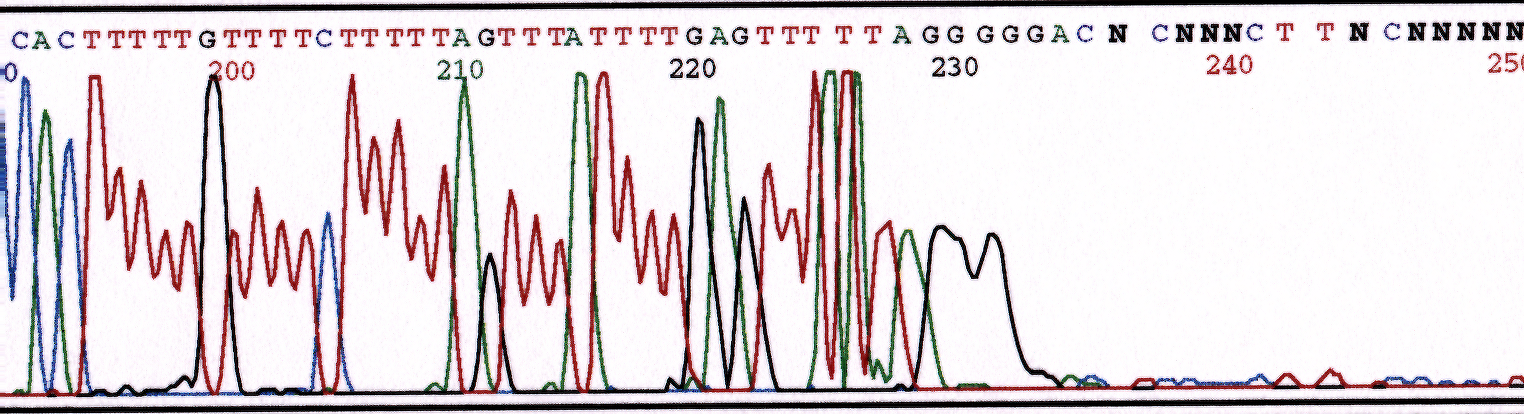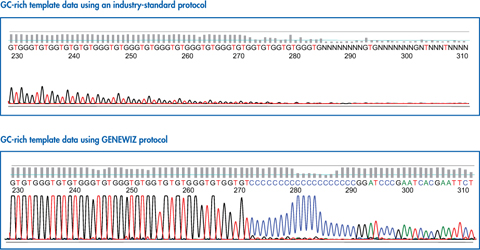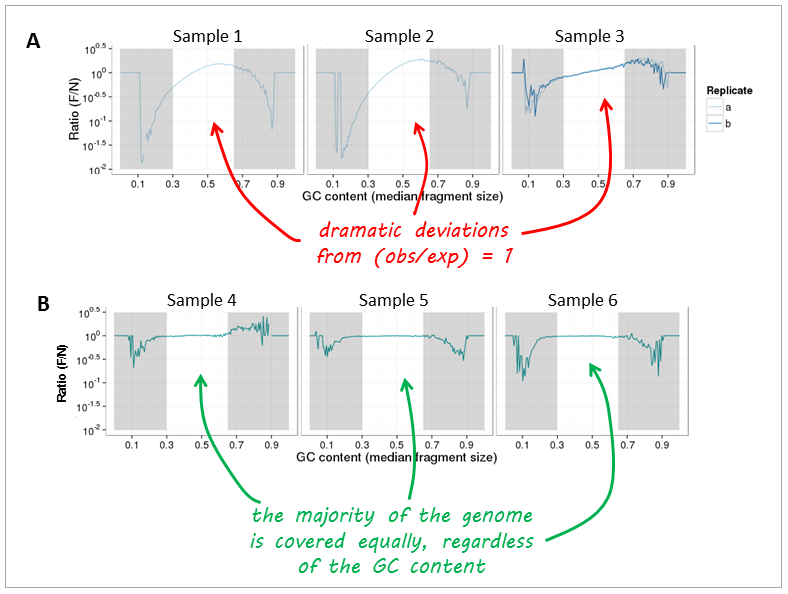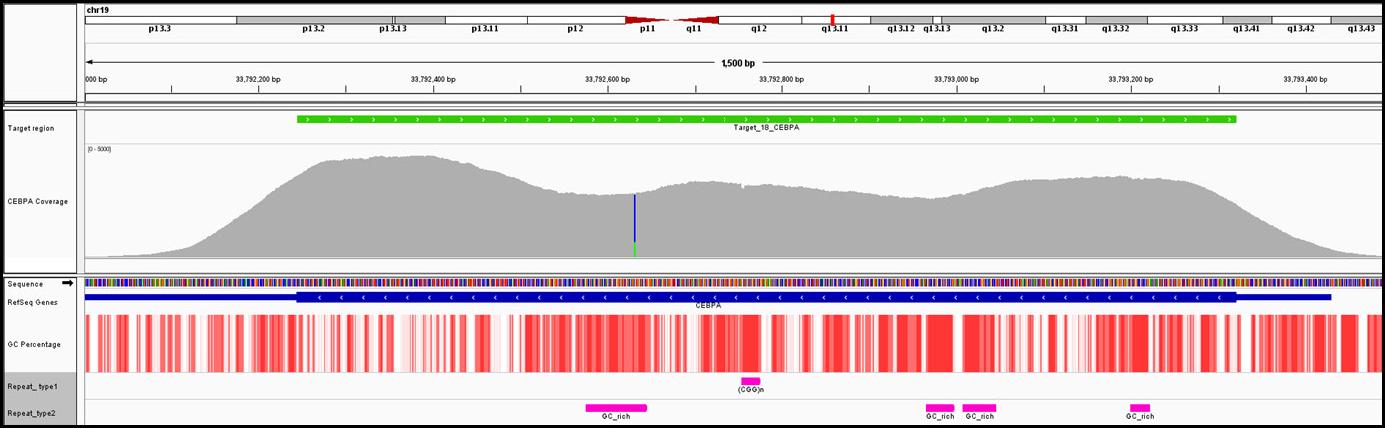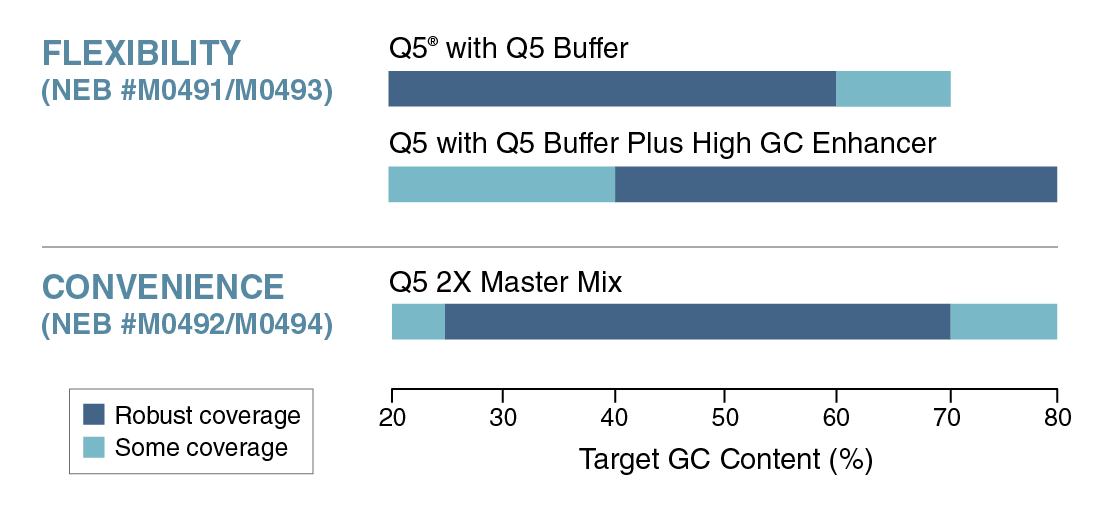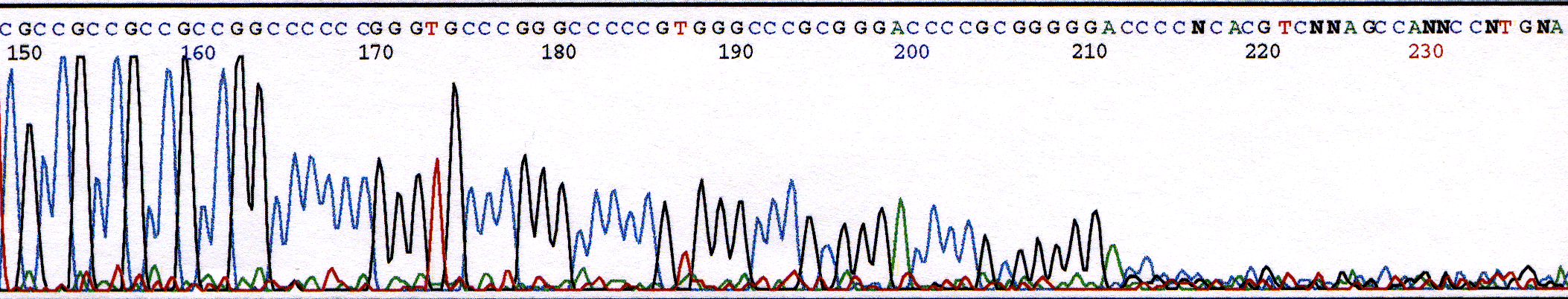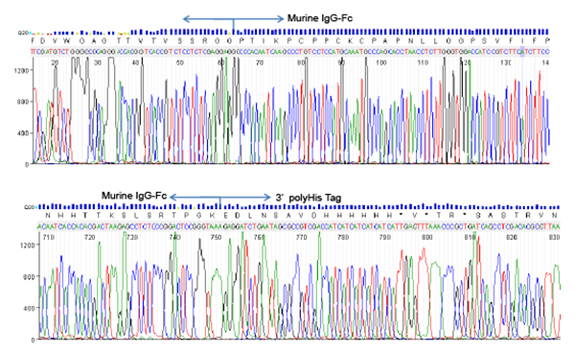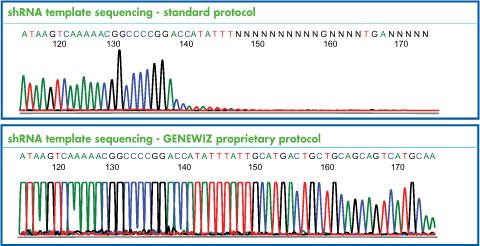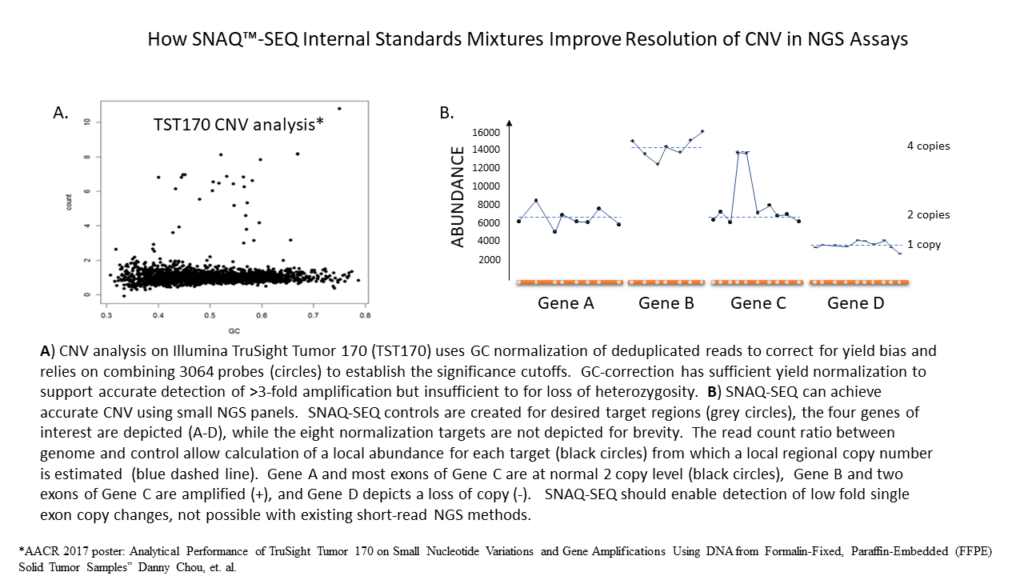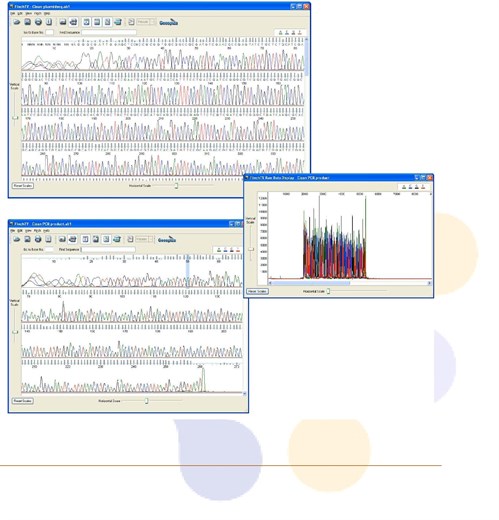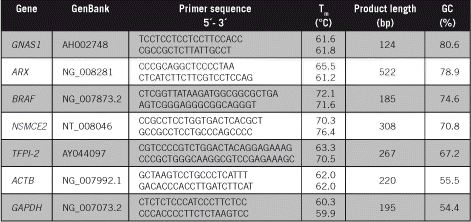
PCR amplification of GC-rich DNA regions using the nucleotide analog N4-methyl-2′-deoxycytidine 5′-triphosphate | BioTechniques

The SSV‐Seq 2.0 PCR‐Free Method Improves the Sequencing of Adeno‐Associated Viral Vector Genomes Containing GC‐Rich Regions and Homopolymers - Lecomte - 2021 - Biotechnology Journal - Wiley Online Library

GC Bias. (A) Schematic of two cycles of GC biased PCR. A sequence with... | Download Scientific Diagram
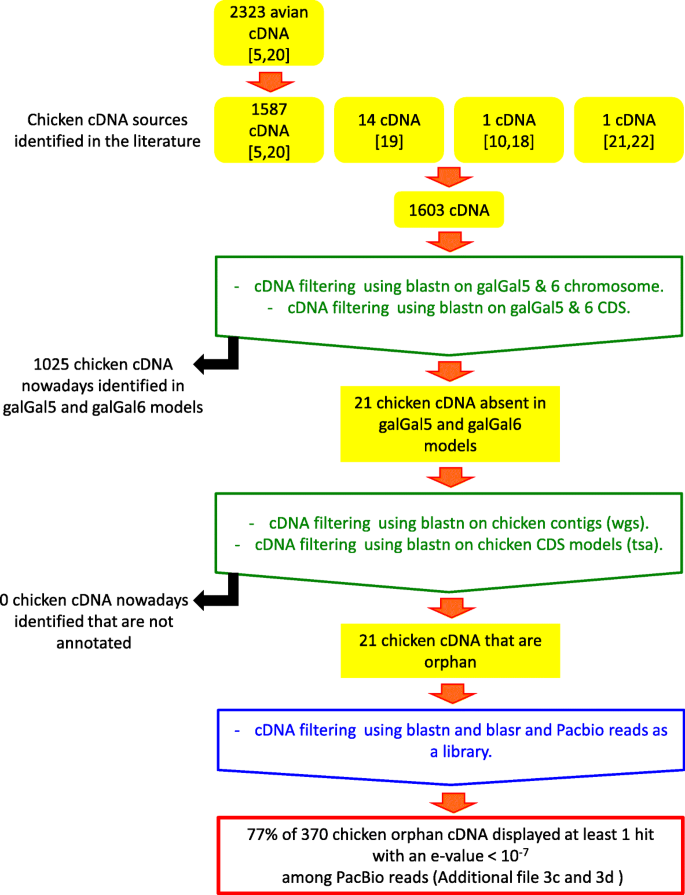
Sequence properties of certain GC rich avian genes, their origins and absence from genome assemblies: case studies | BMC Genomics | Full Text
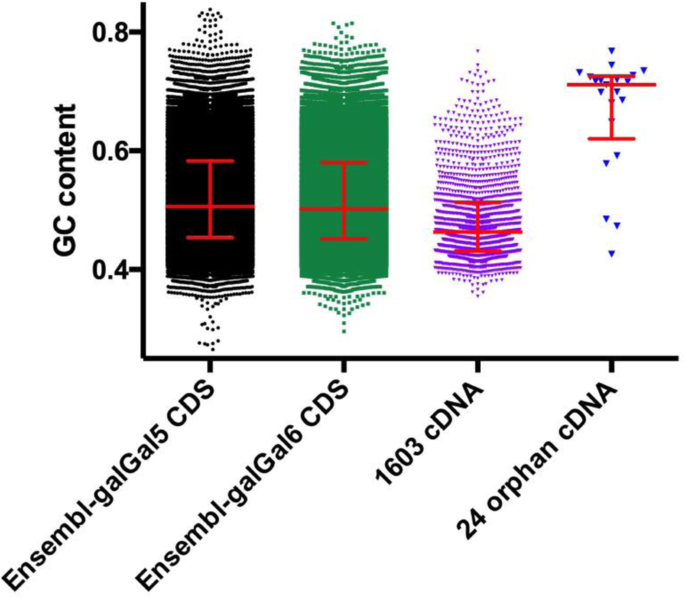
Sequence properties of certain GC rich avian genes, their origins and absence from genome assemblies: case studies | BMC Genomics | Full Text
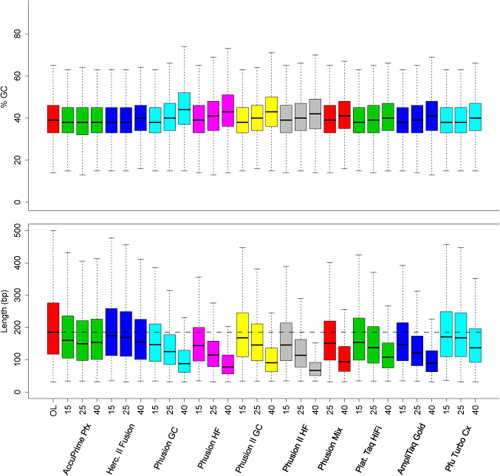
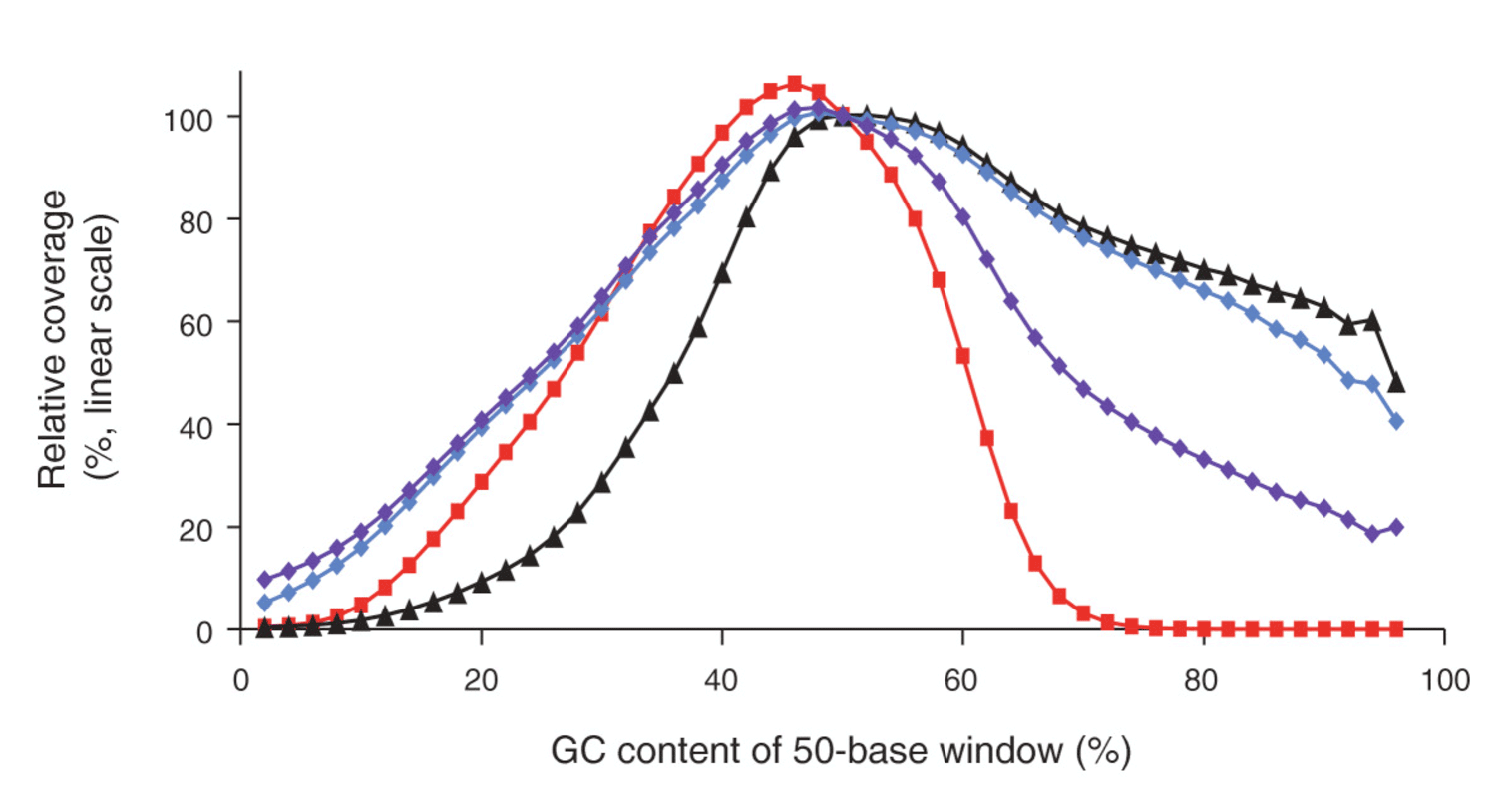
Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries | BioTechniques