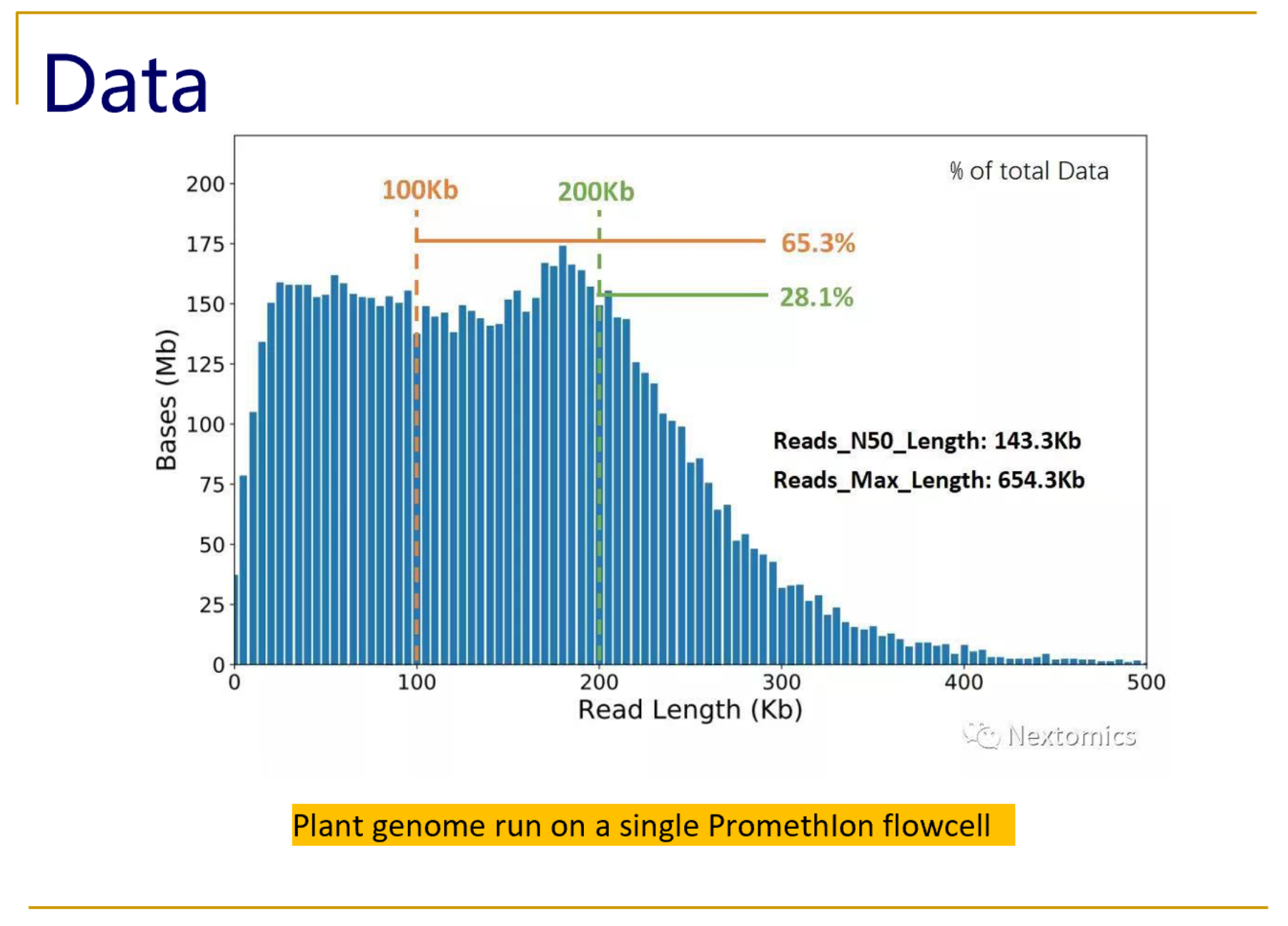
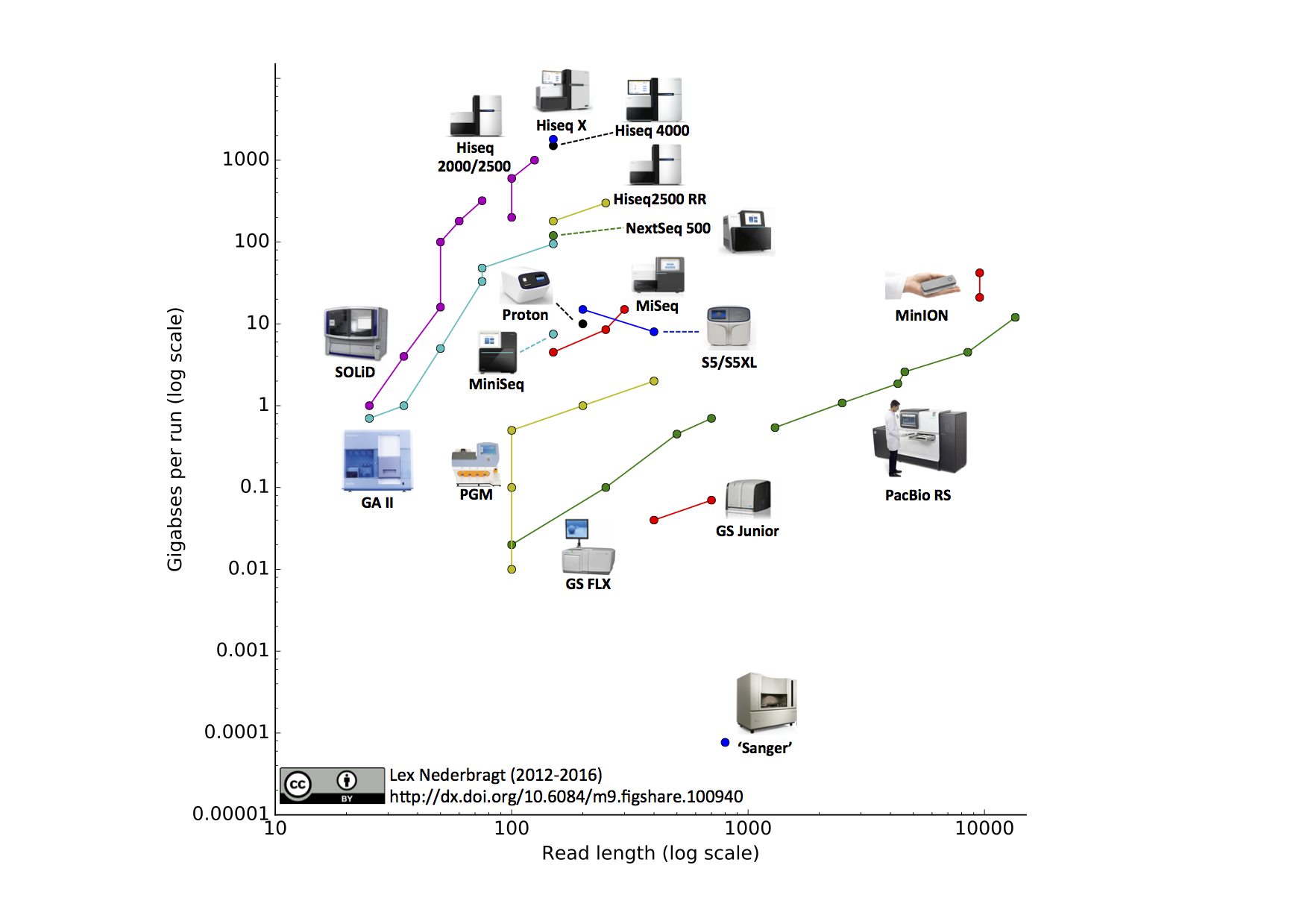
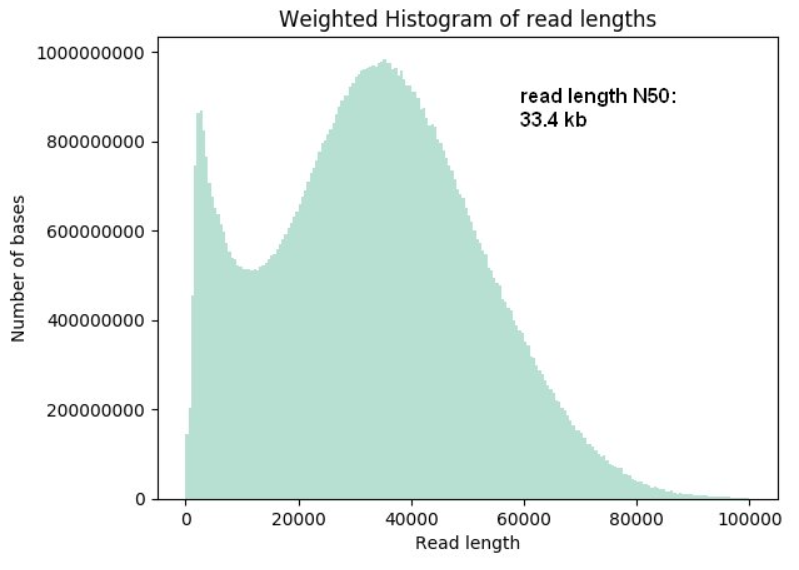
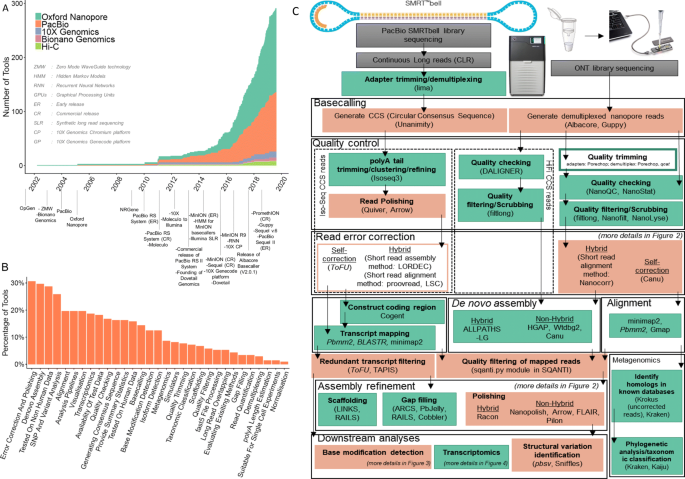
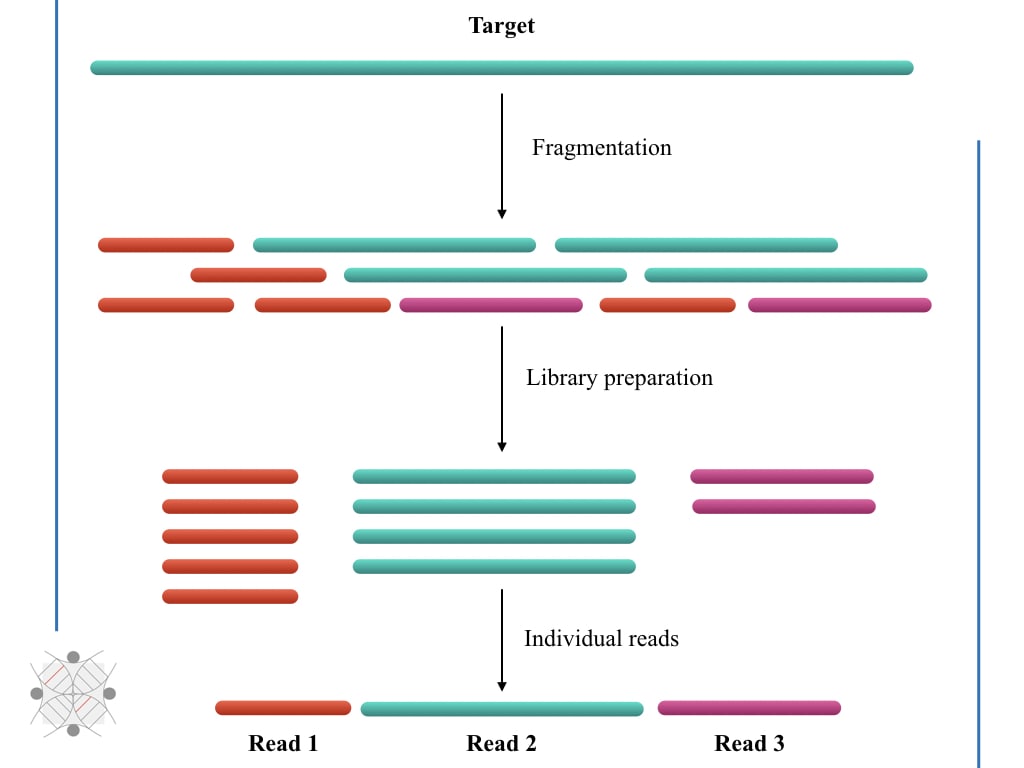
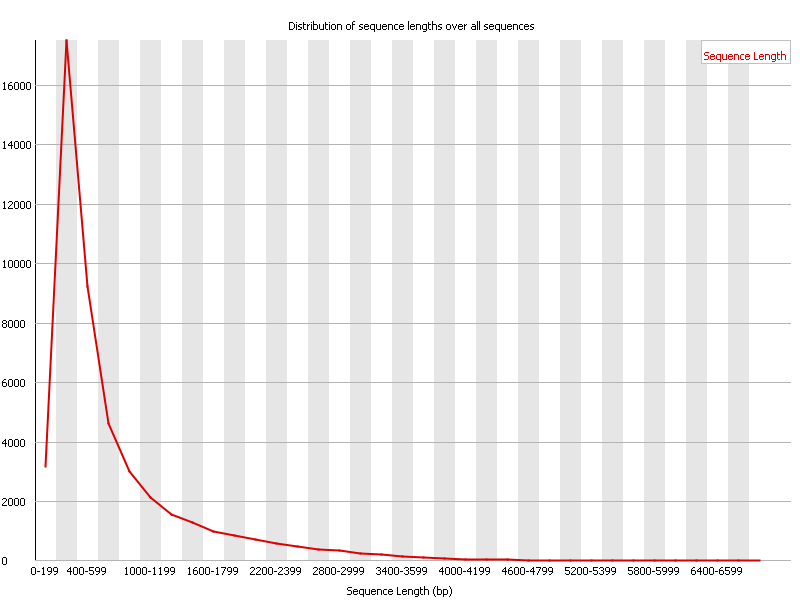
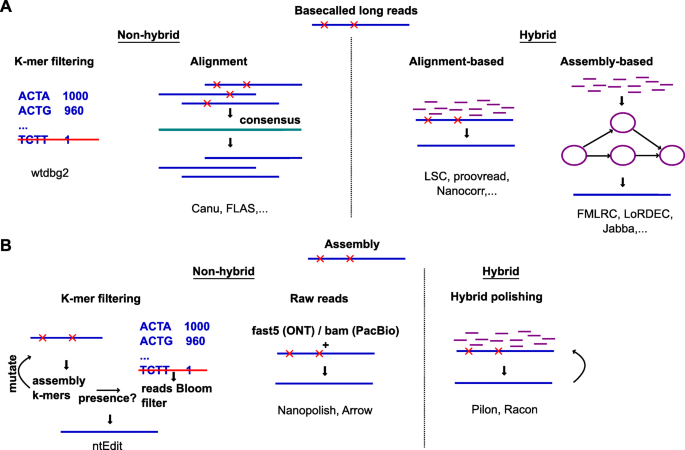
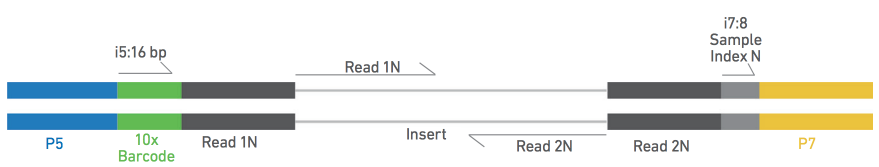
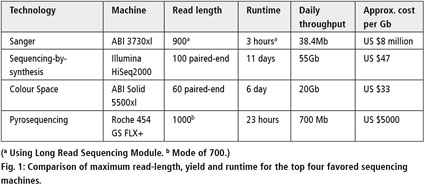
Direct Chloroplast Sequencing: Comparison of Sequencing Platforms and Analysis Tools for Whole Chloroplast Barcoding | PLOS ONE
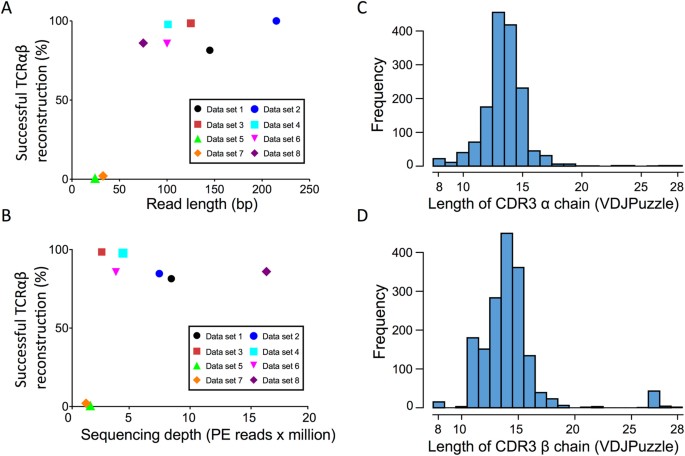
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

Longer and longer: DNA sequence of more than two million bases now achieved with nanopore sequencing.

Oxford Nanopore Technology: A Promising Long-Read Sequencing Platform To Study Exon Connectivity and Characterize Isoforms of Complex Genes | Semantic Scholar
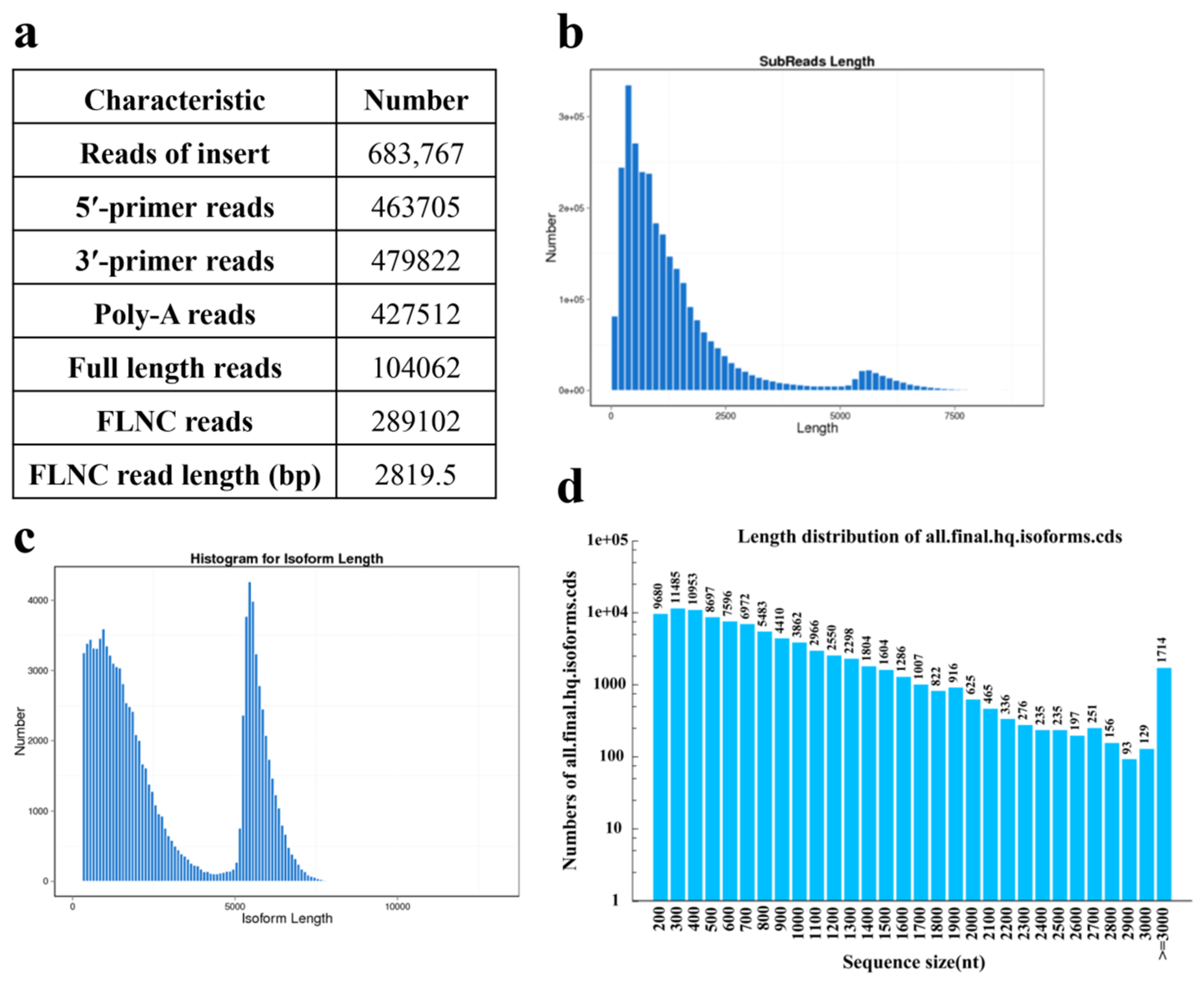
IJMS | Free Full-Text | Full-Length Transcriptome Sequencing and Different Chemotype Expression Profile Analysis of Genes Related to Monoterpenoid Biosynthesis in Cinnamomum porrectum