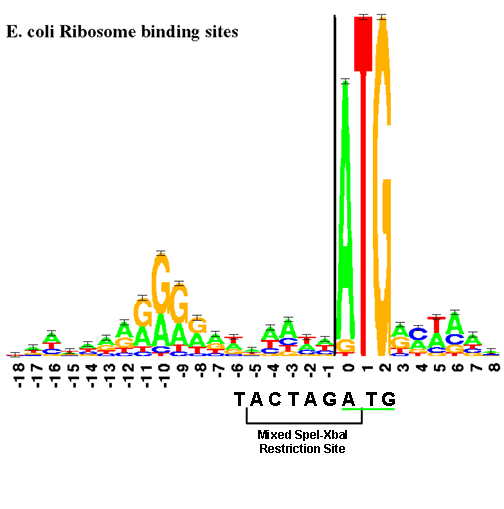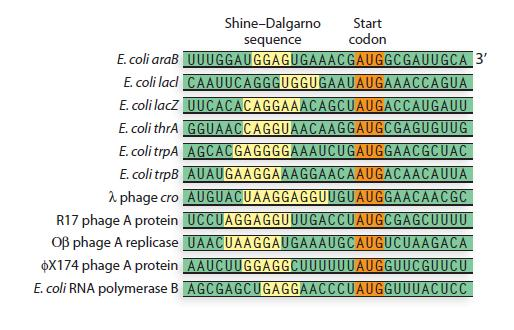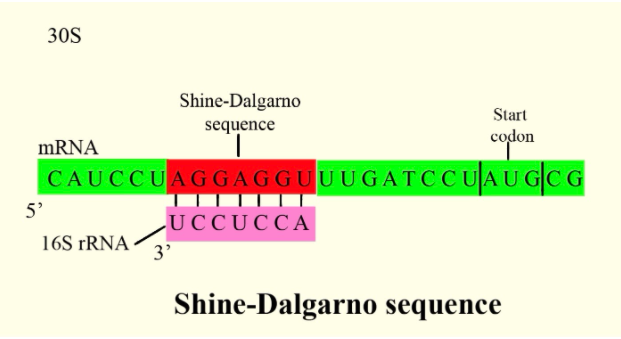
In prokaryotes, the ribosomes binding site on mRNA is called(a)Hogness sequence(b)Shine- Dalgarno sequence(c)Prinbow sequence(d)TATA- box

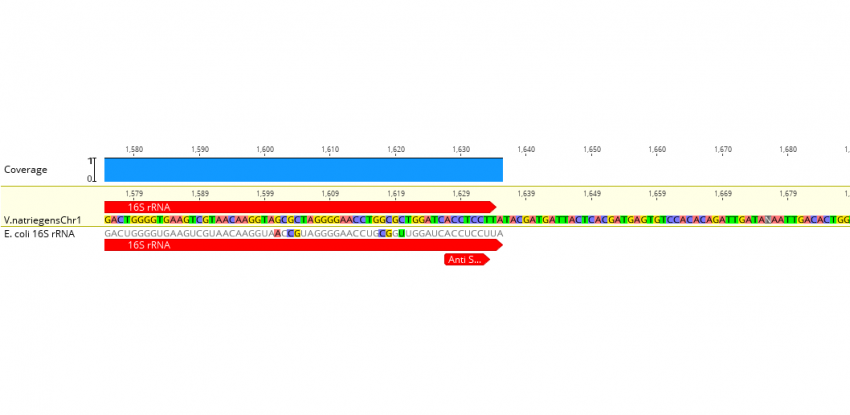
Nucleotide sequence of the putative anti-Shine–Dalgarno sequence from... | Download Scientific Diagram
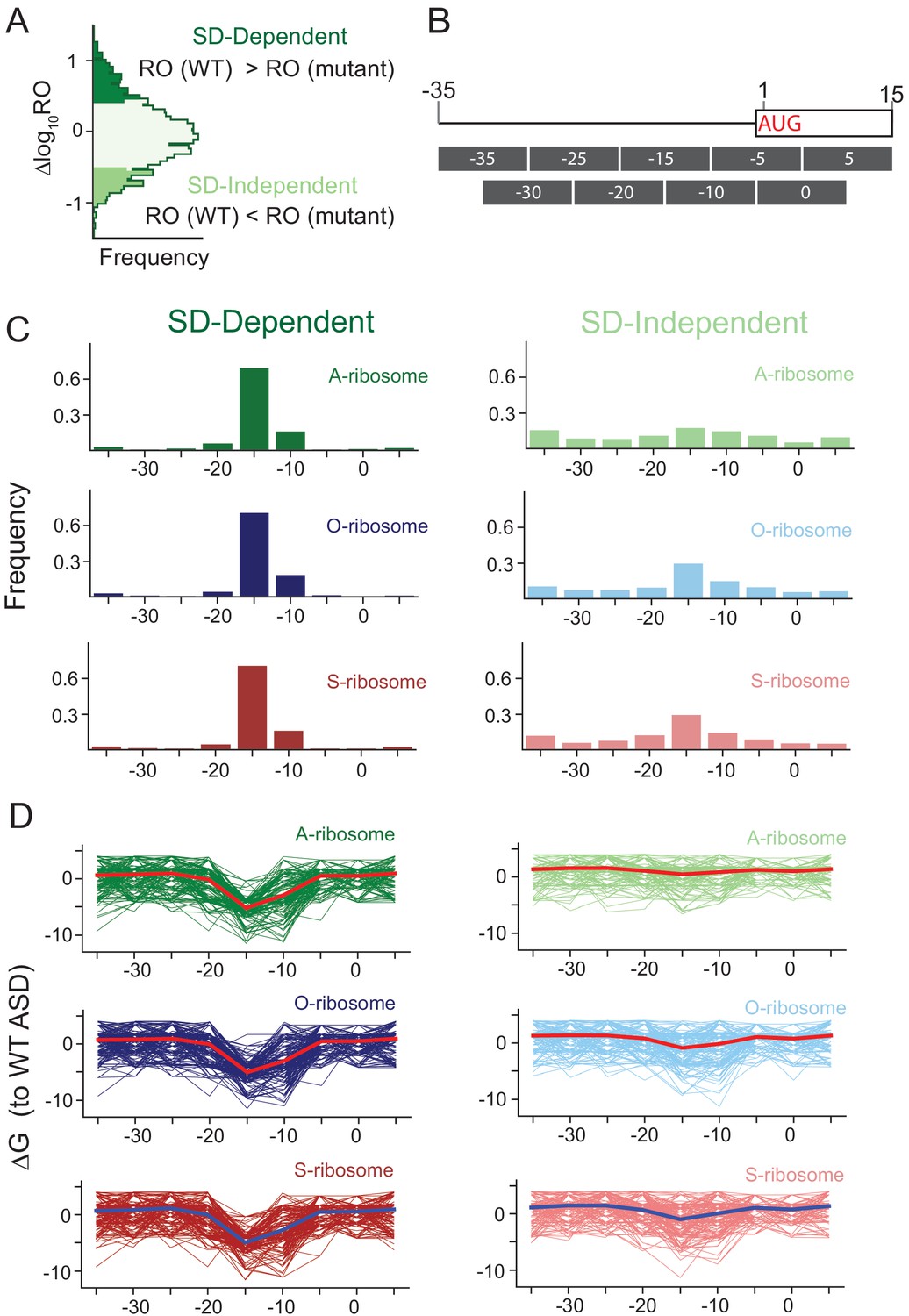
Defining the anti-Shine-Dalgarno sequence interaction and quantifying its functional role in regulating translation efficiency

The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts | Nature Communications

The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis | Microbial Cell Factories | Full Text

Accessibility of the Shine-Dalgarno Sequence Dictates N-Terminal Codon Bias in E. coli - ScienceDirect

What is the Difference Between Shine Dalgarno and Kozak Sequence | Compare the Difference Between Similar Terms

Diversity of translation initiation mechanisms across bacterial species is driven by environmental conditions and growth demands | bioRxiv
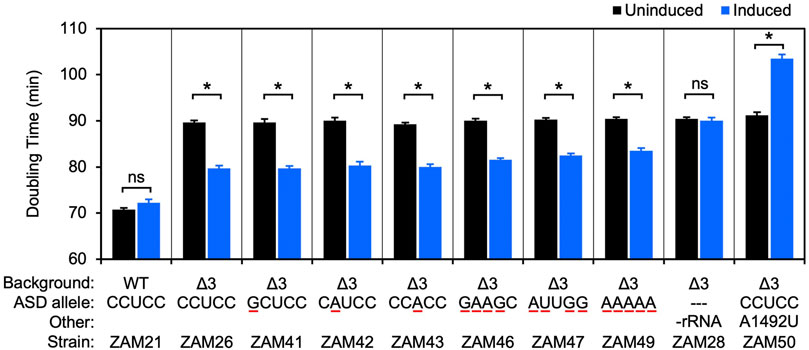
Frontiers | Comparative Analysis of anti-Shine- Dalgarno Function in Flavobacterium johnsoniae and Escherichia coli

Influences on gene expression in vivo by a Shine–Dalgarno sequence - Jin - 2006 - Molecular Microbiology - Wiley Online Library
Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency - Publications - Amaral Lab
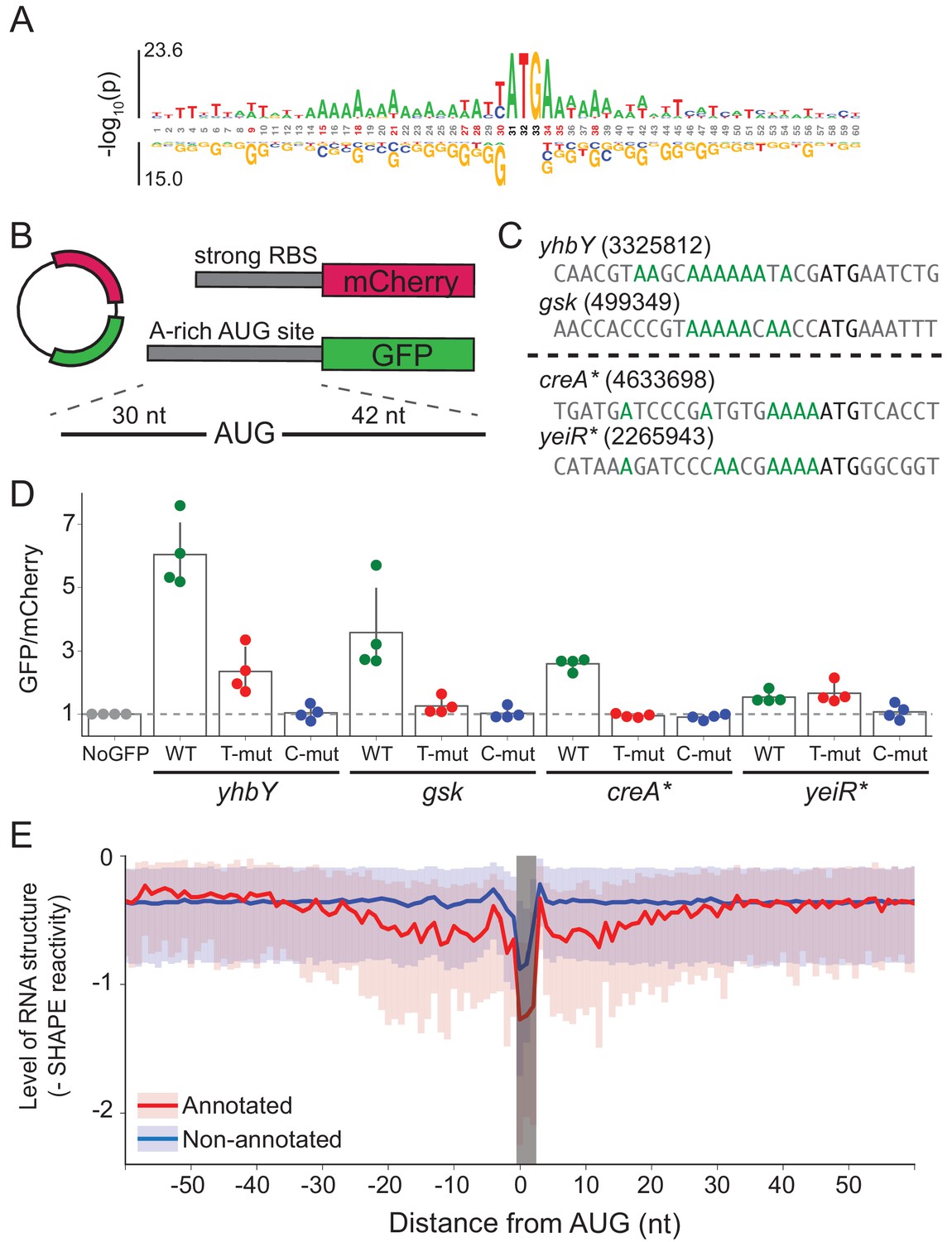
Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing | eLife

Influences on gene expression in vivo by a Shine–Dalgarno sequence - Jin - 2006 - Molecular Microbiology - Wiley Online Library

Silent mutations in the Shine-Dalgarno-like sequence and the predicted... | Download Scientific Diagram

Multiple clustalW alignment of-35 box,-10 box, and Shine-Dalgarno (SD)... | Download Scientific Diagram

Influence of the spacer region between the Shine–Dalgarno box and the start codon for fine‐tuning of the translation efficiency in Escherichia coli - Komarova - 2020 - Microbial Biotechnology - Wiley Online Library