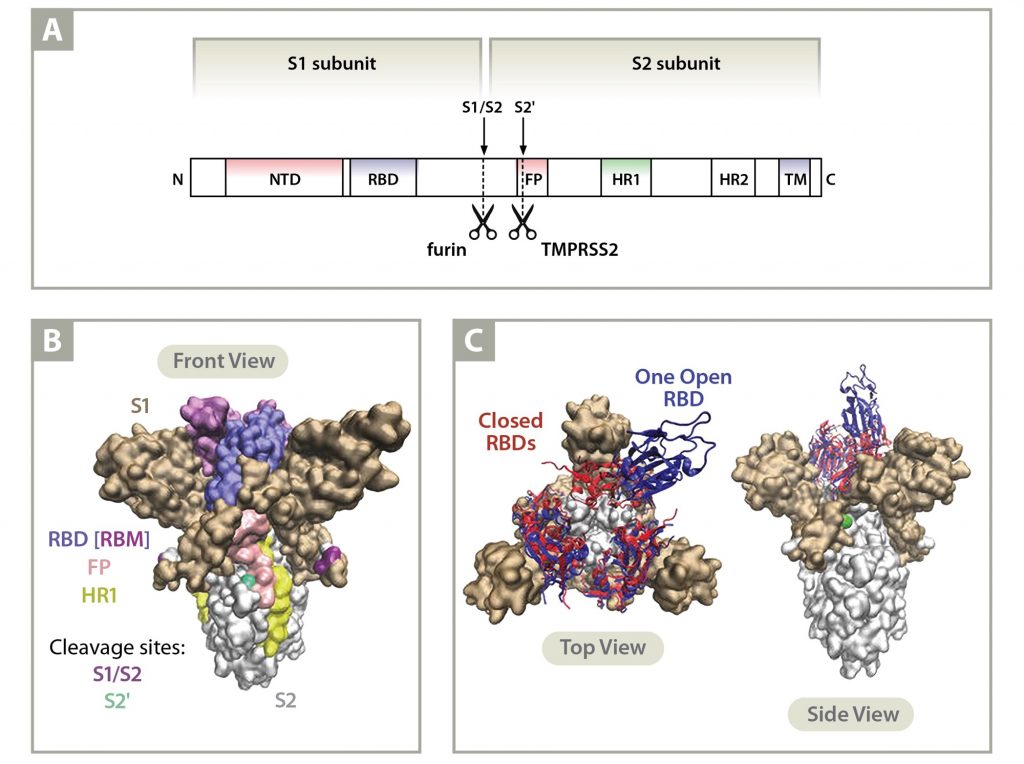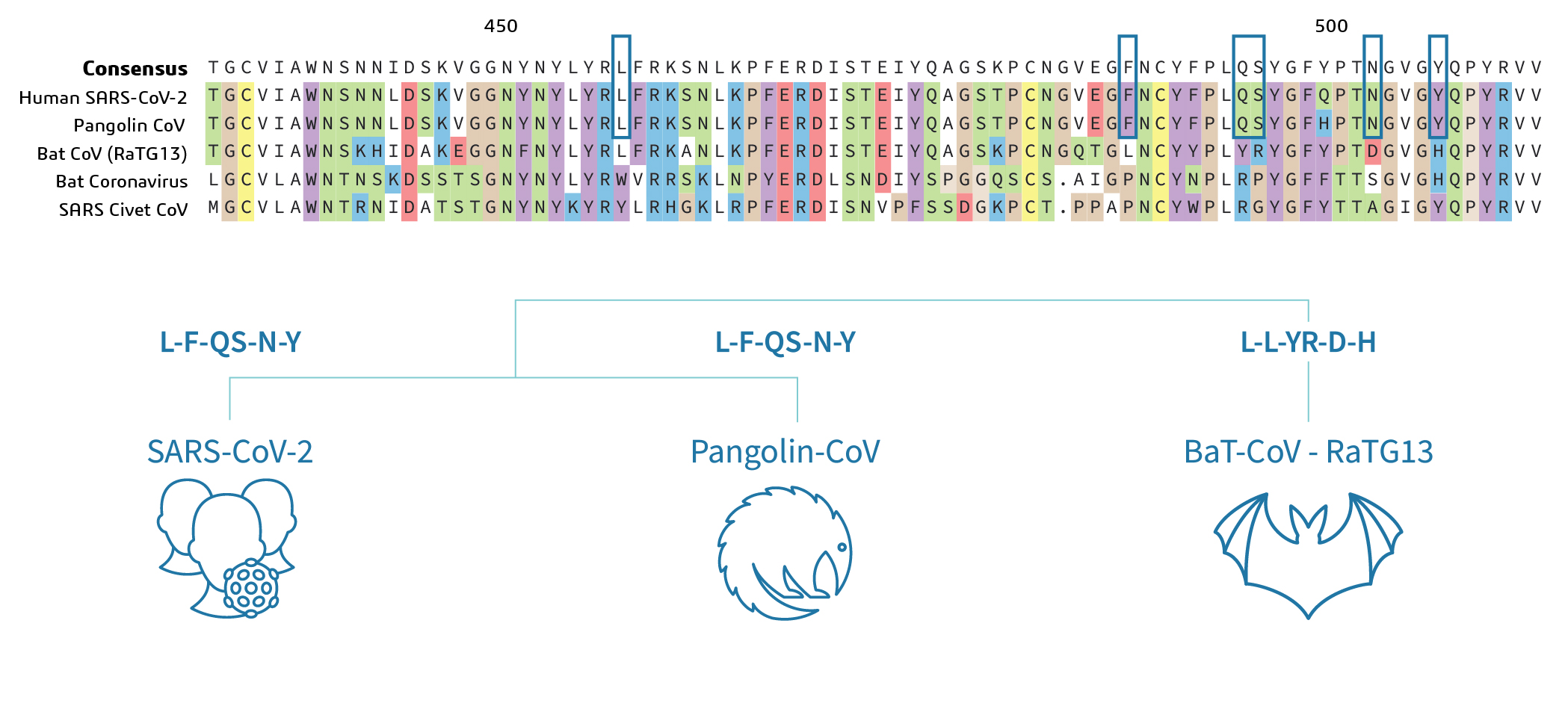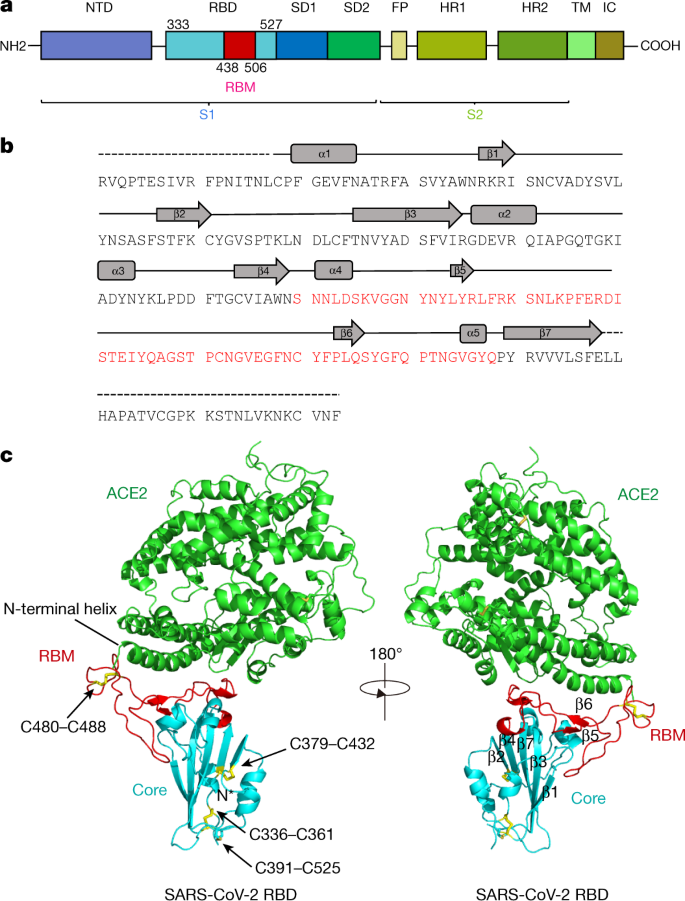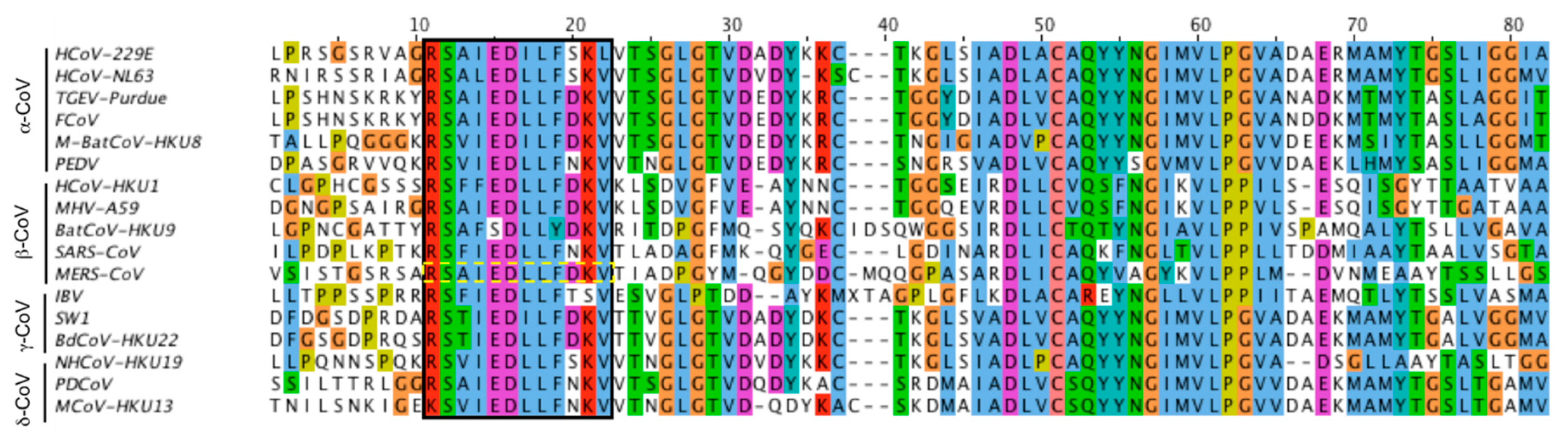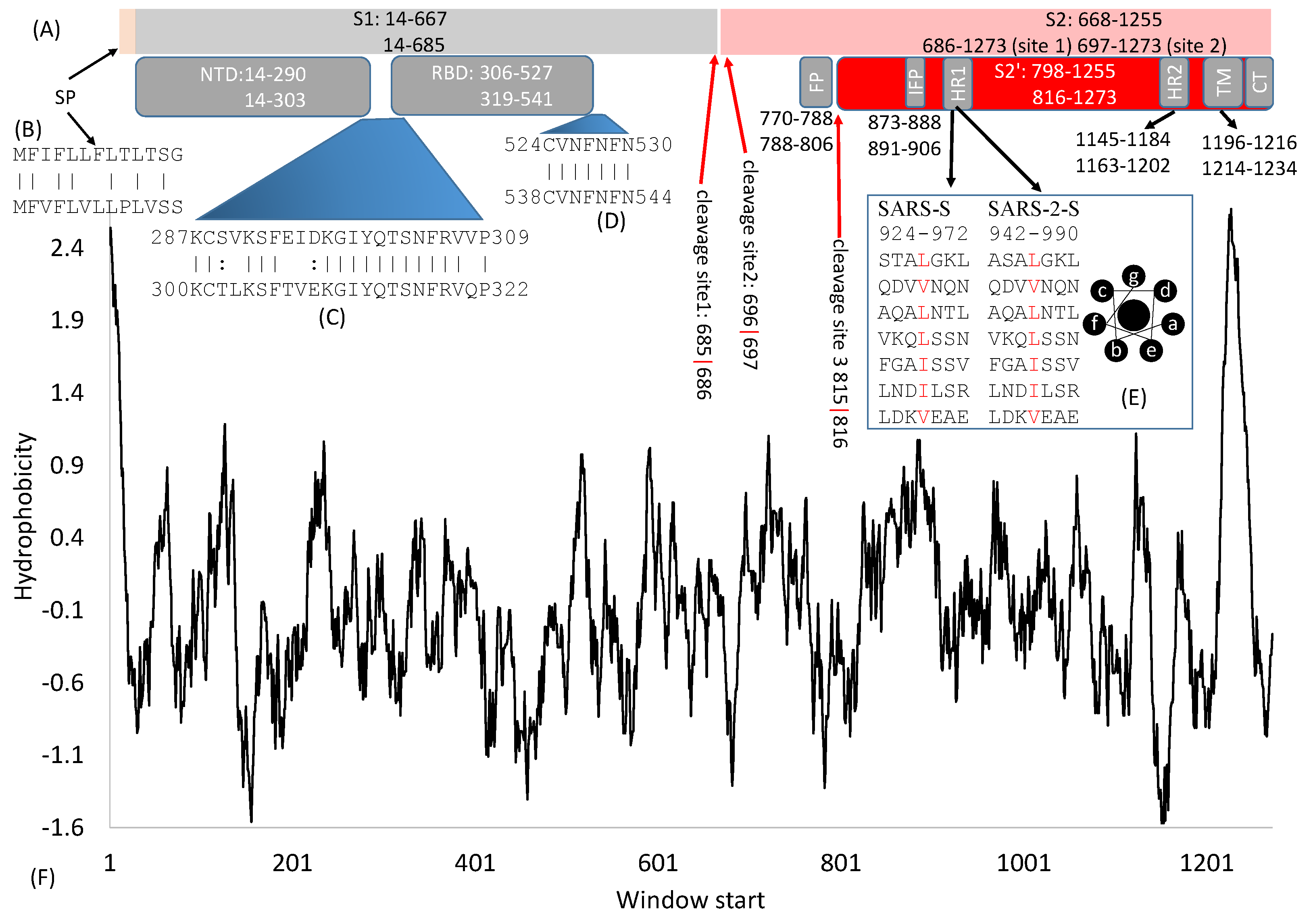
Viruses | Free Full-Text | Domains and Functions of Spike Protein in SARS-Cov-2 in the Context of Vaccine Design

Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein | Journal of Clinical Pathology

Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity - eBioMedicine

Spike protein sequences of Cambodian, Thai and Japanese bat sarbecoviruses provide insights into the natural evolution of the Receptor Binding Domain and S1/S2 cleavage site - Virological
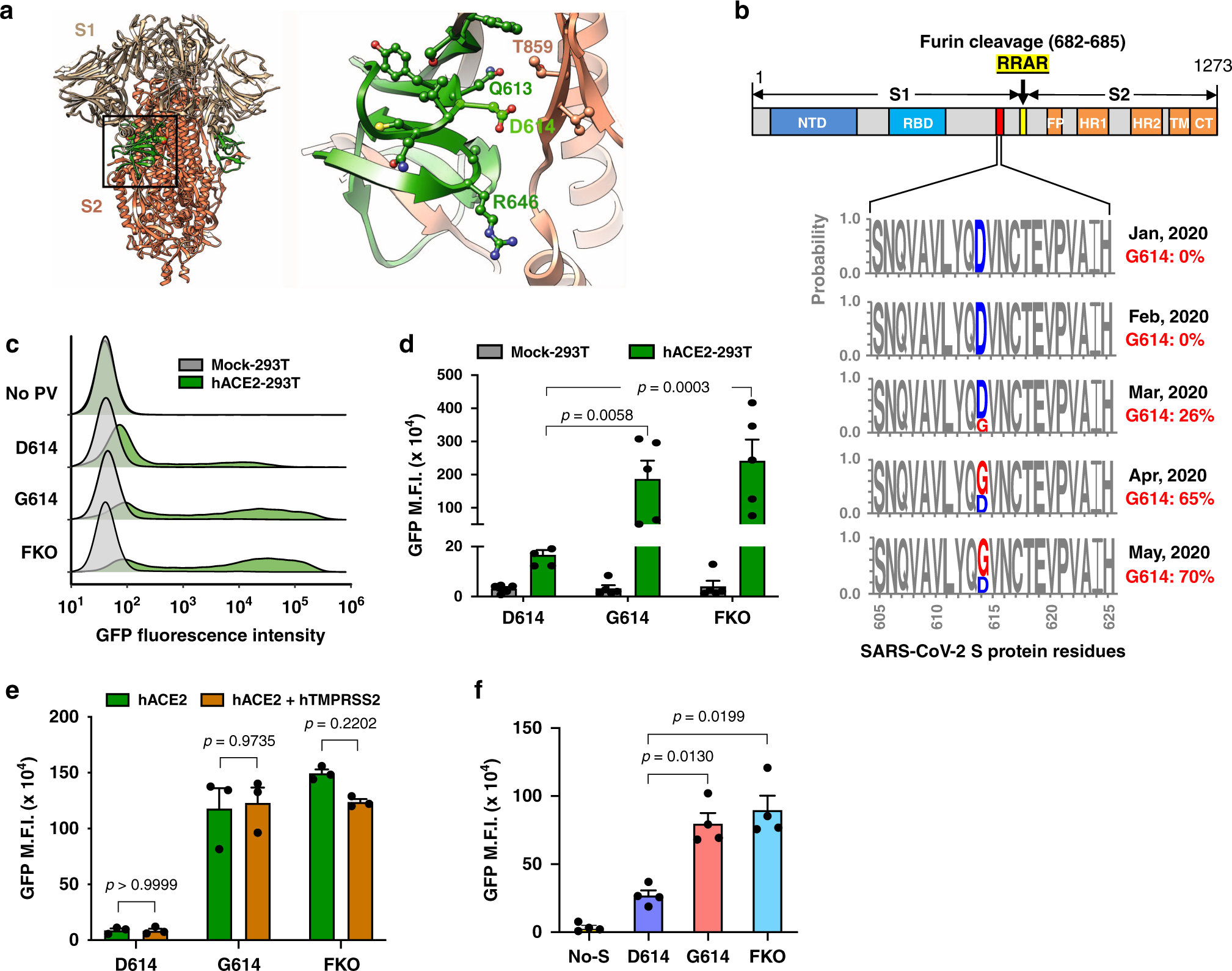
SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity | Nature Communications
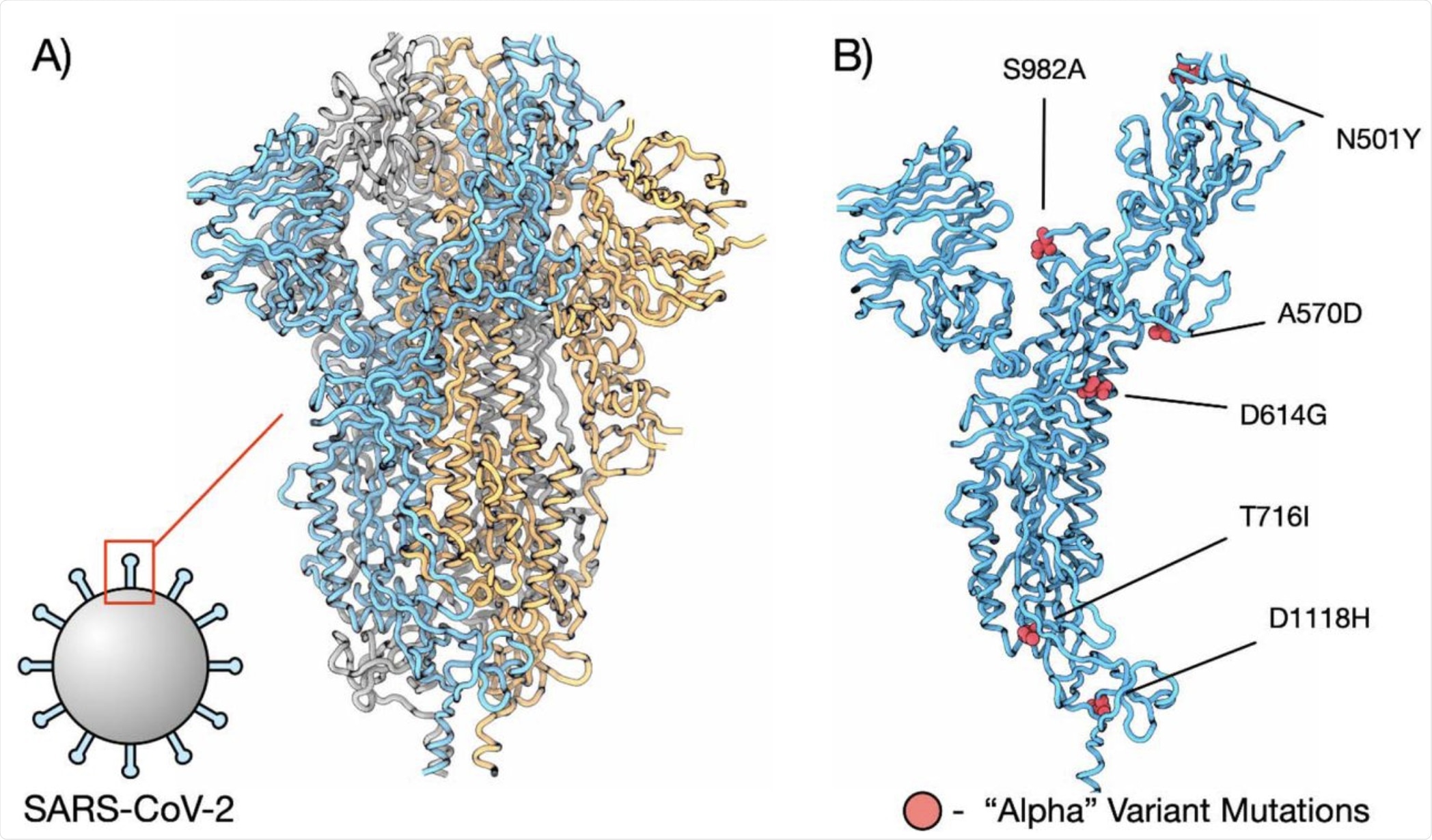
Mutations in the spike proteins of SARS-CoV-2 select for amino acid changes, increasing protein stability

SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex | Science
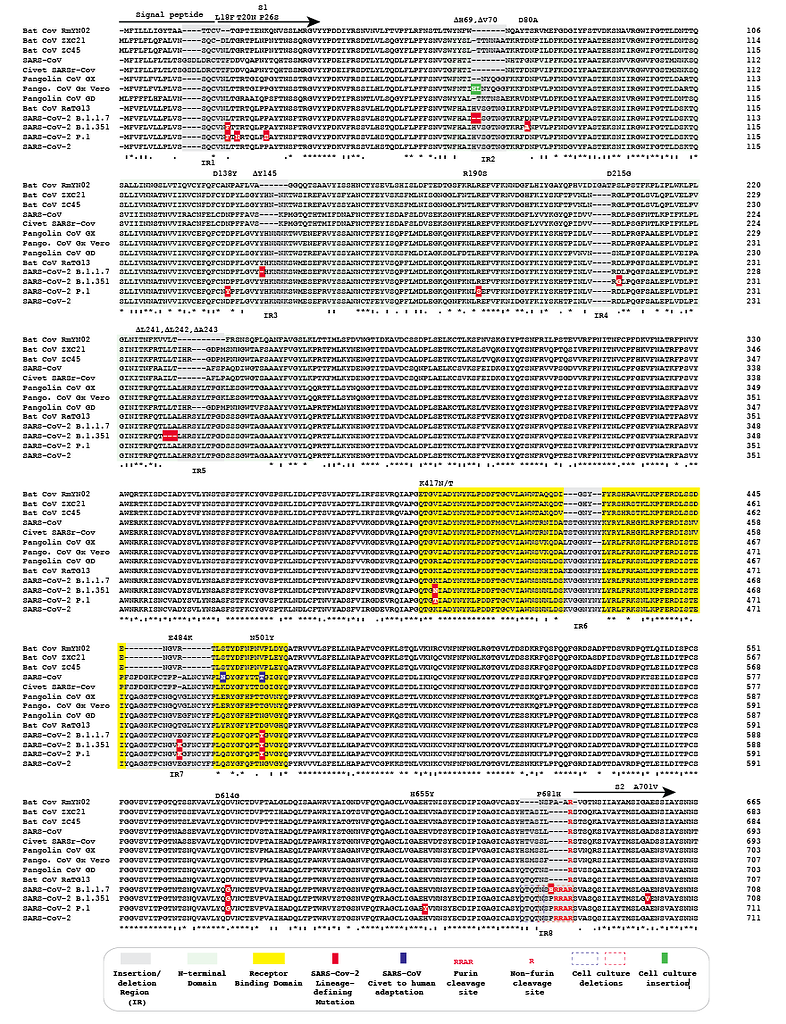
Spike protein mutations in novel SARS-CoV-2 'variants of concern' commonly occur in or near indels - Virological

Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts | Journal of Virology

Newcastle disease virus (NDV) expressing the spike protein of SARS-CoV-2 as a live virus vaccine candidate - eBioMedicine

Tracking the amino acid changes of spike proteins across diverse host species of severe acute respiratory syndrome coronavirus 2 - ScienceDirect

Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: a computational biology approach | bioRxiv