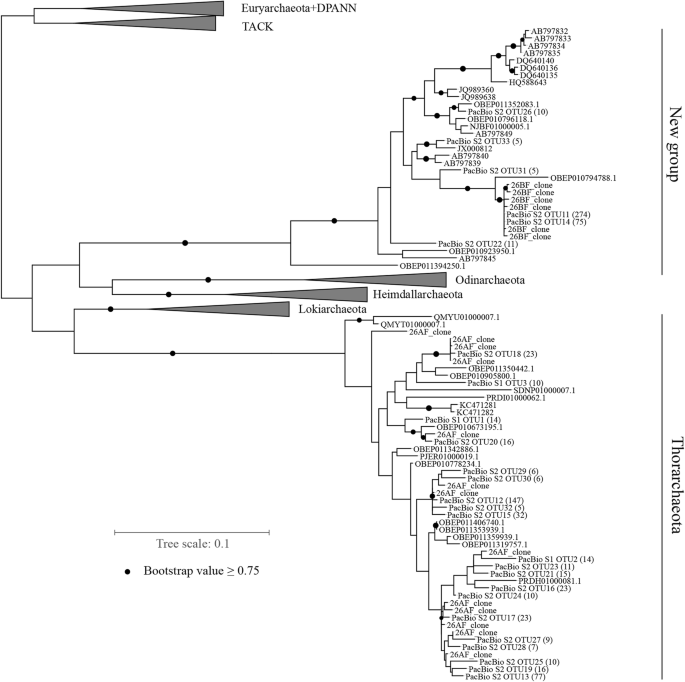
Characterising the Canine Oral Microbiome by Direct Sequencing of Reverse-Transcribed rRNA Molecules | PLOS ONE
Fact Sheet: rRNA in Evolutionary Studies and Environmental Sampling – microBEnet: the microbiology of the Built Environment network

Sequence-Specific Cleavage of Small-Subunit (SSU) rRNA with Oligonucleotides and RNase H: a Rapid and Simple Approach to SSU rRNA-Based Quantitative Detection of Microorganisms | Applied and Environmental Microbiology
Modified RNA-seq method for microbial community and diversity analysis using rRNA in different types of environmental samples | PLOS ONE
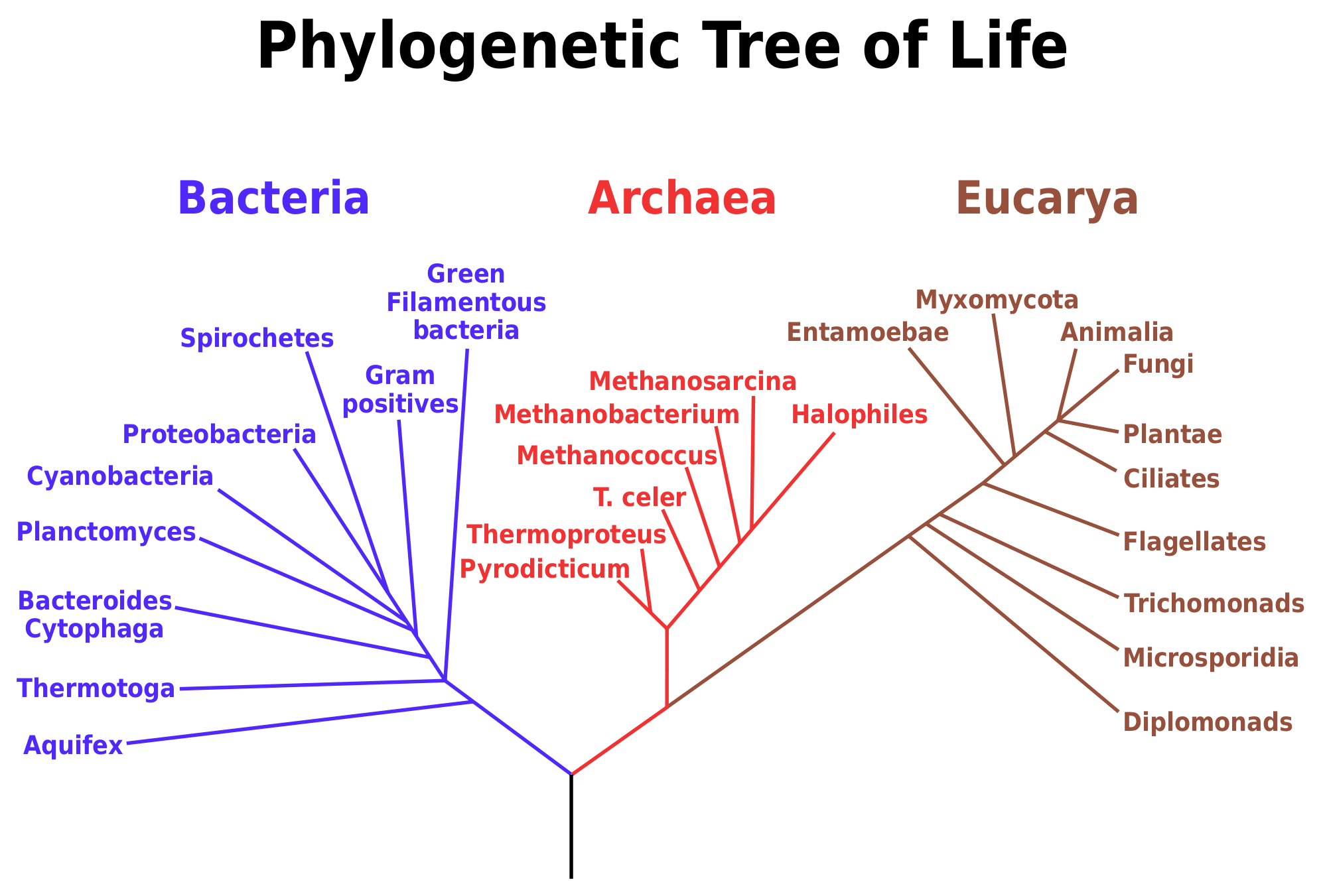
Universal phylogenetic tree based on comparison of SSU rRNA sequences.... | Download Scientific Diagram
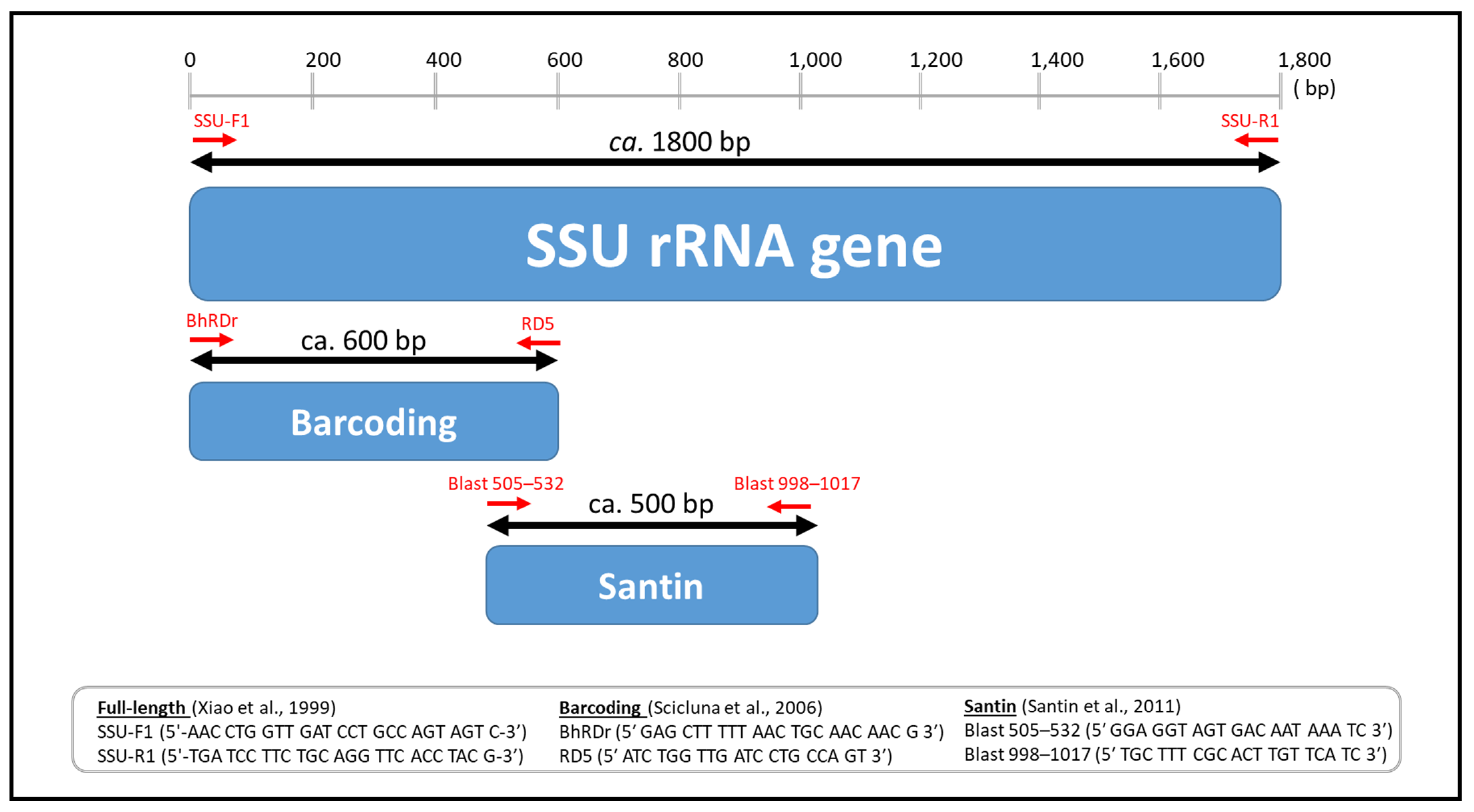
Microorganisms | Free Full-Text | Mind the Gap: New Full-Length Sequences of Blastocystis Subtypes Generated via Oxford Nanopore Minion Sequencing Allow for Comparisons between Full-Length and Partial Sequences of the Small Subunit

Impact of 16S rRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases | Clinical Microbiology Reviews
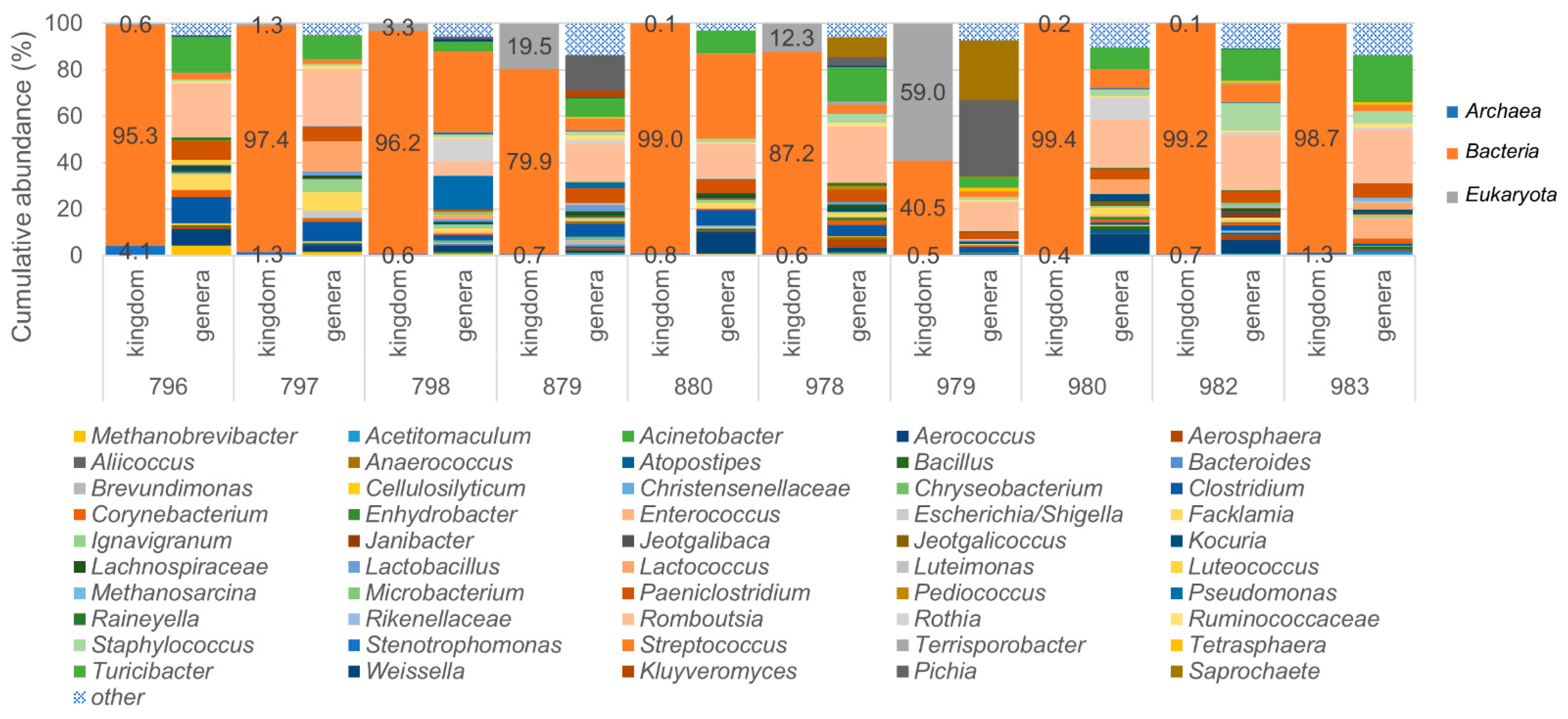
Microorganisms | Free Full-Text | Full-Length SSU rRNA Gene Sequencing Allows Species-Level Detection of Bacteria, Archaea, and Yeasts Present in Milk
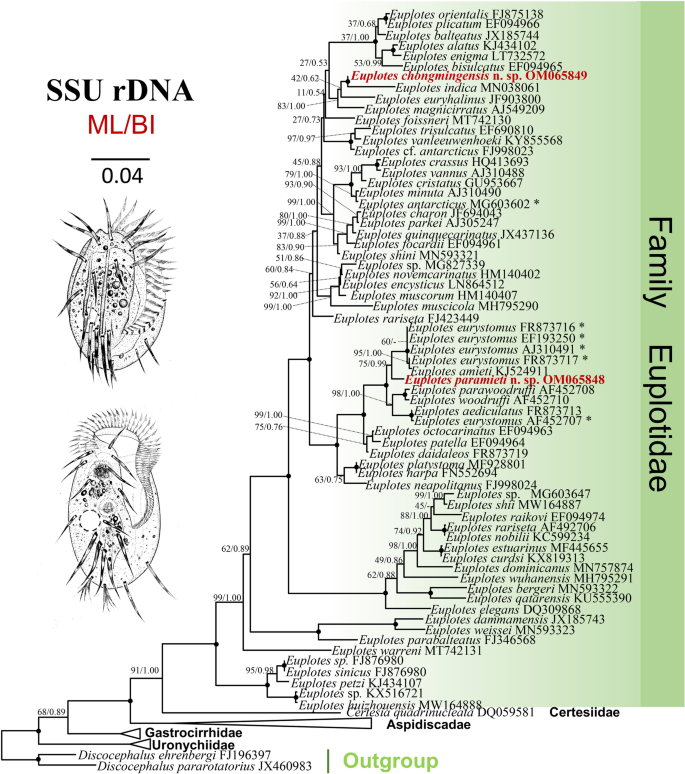
Taxonomy and SSU rRNA gene-based phylogeny of two new Euplotes species from China: E. chongmingensis n. sp. and E. paramieti n. sp. (Protista, Ciliophora) | BMC Microbiology | Full Text

Does Intraspecific Variation in rDNA Copy Number Affect Analysis of Microbial Communities?: Trends in Microbiology

Disentangling sources of variation in SSU rDNA sequences from single cell analyses of ciliates: impact of copy number variation and experimental error | Proceedings of the Royal Society B: Biological Sciences
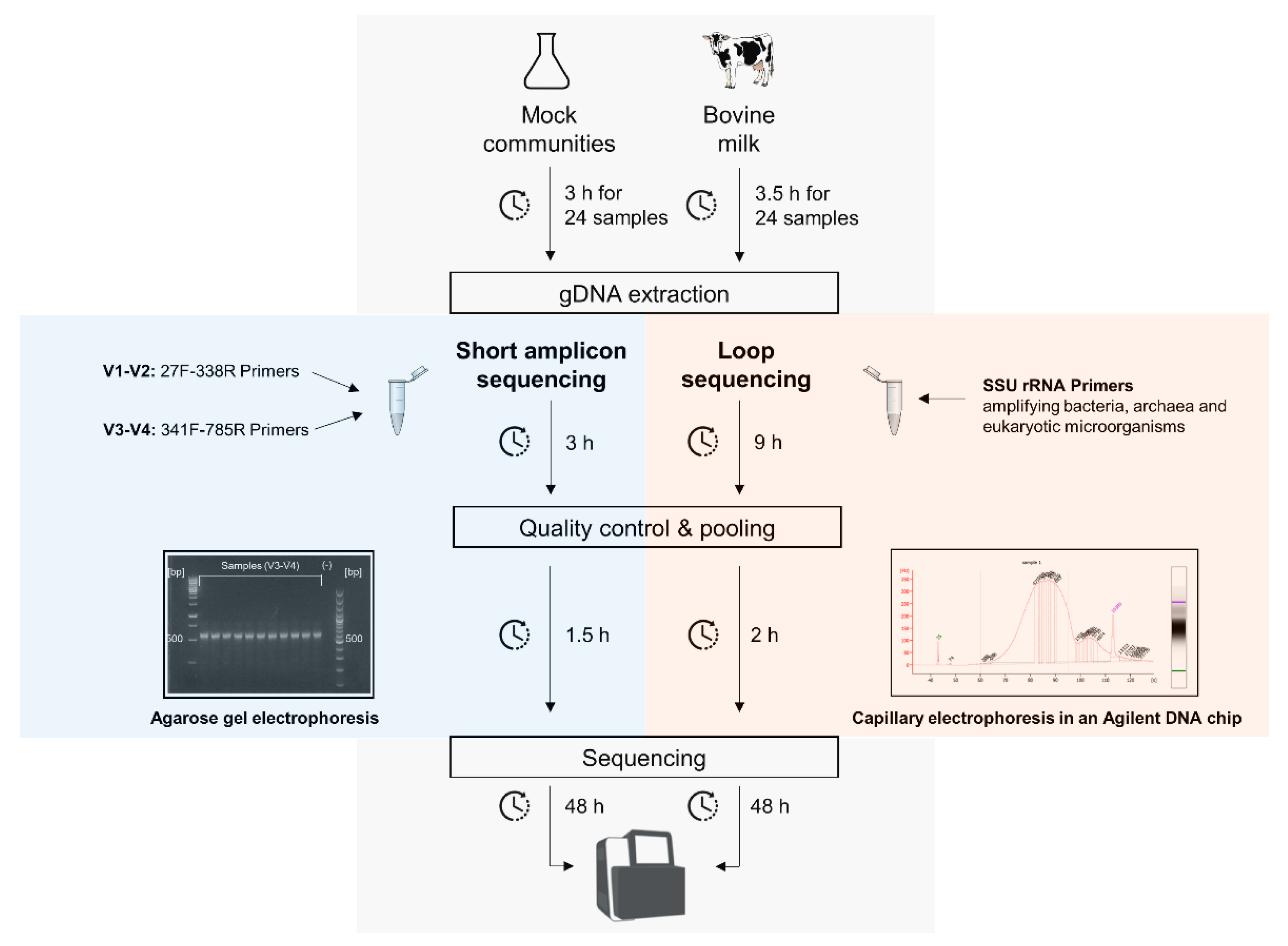
Microorganisms | Free Full-Text | Full-Length SSU rRNA Gene Sequencing Allows Species-Level Detection of Bacteria, Archaea, and Yeasts Present in Milk

Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota | Scientific Reports
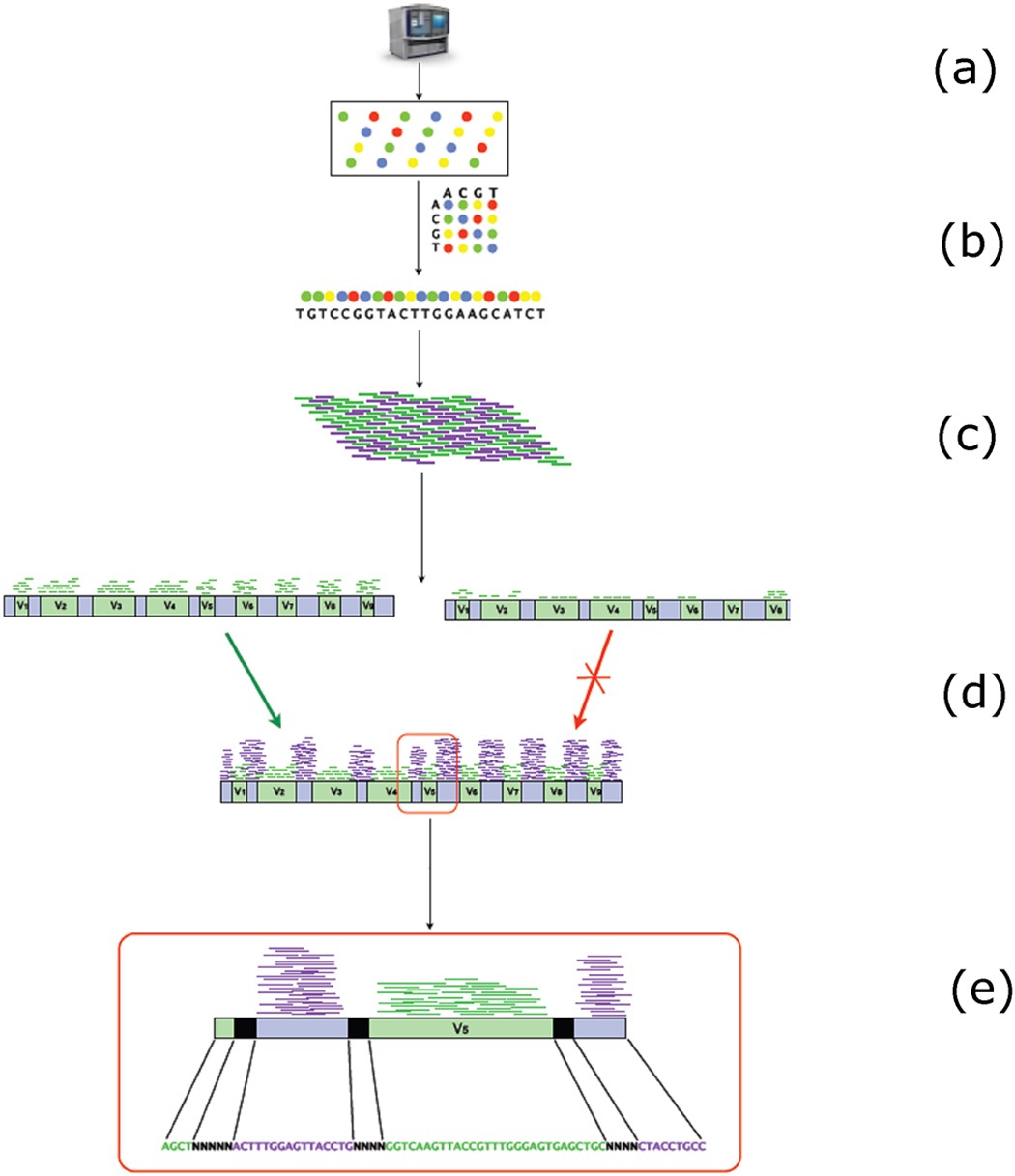
Direct 16S rRNA-seq from bacterial communities: a PCR-independent approach to simultaneously assess microbial diversity and functional activity potential of each taxon | Scientific Reports
![PDF] A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq | Semantic Scholar PDF] A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/c5433f6643987939f74806d2f91db31fe7eac03a/4-Figure1-1.png)
PDF] A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq | Semantic Scholar