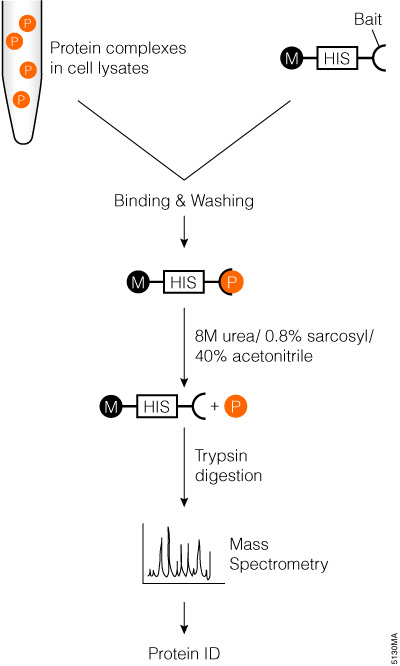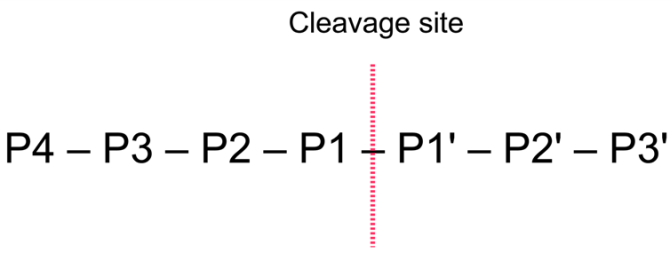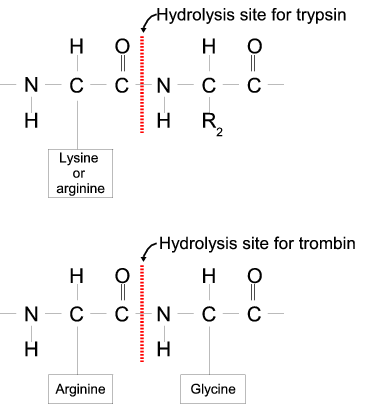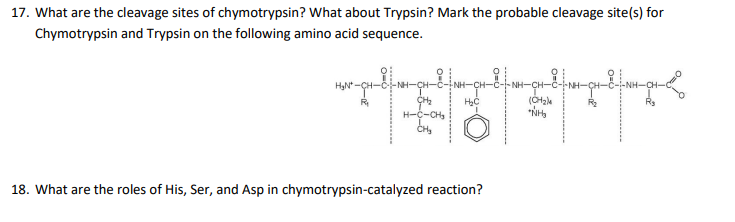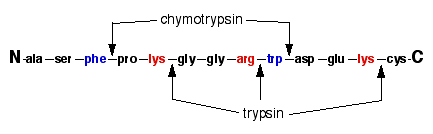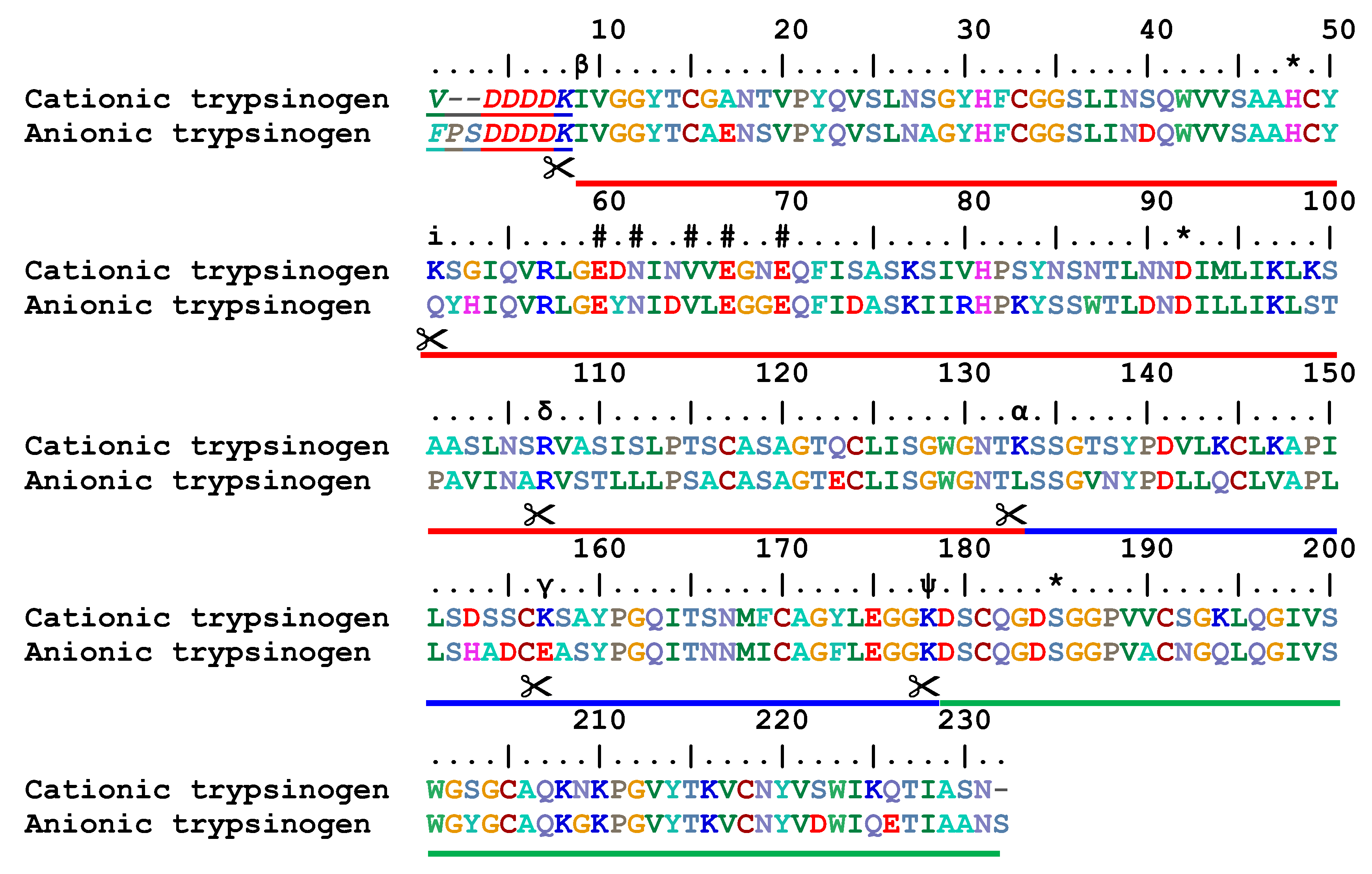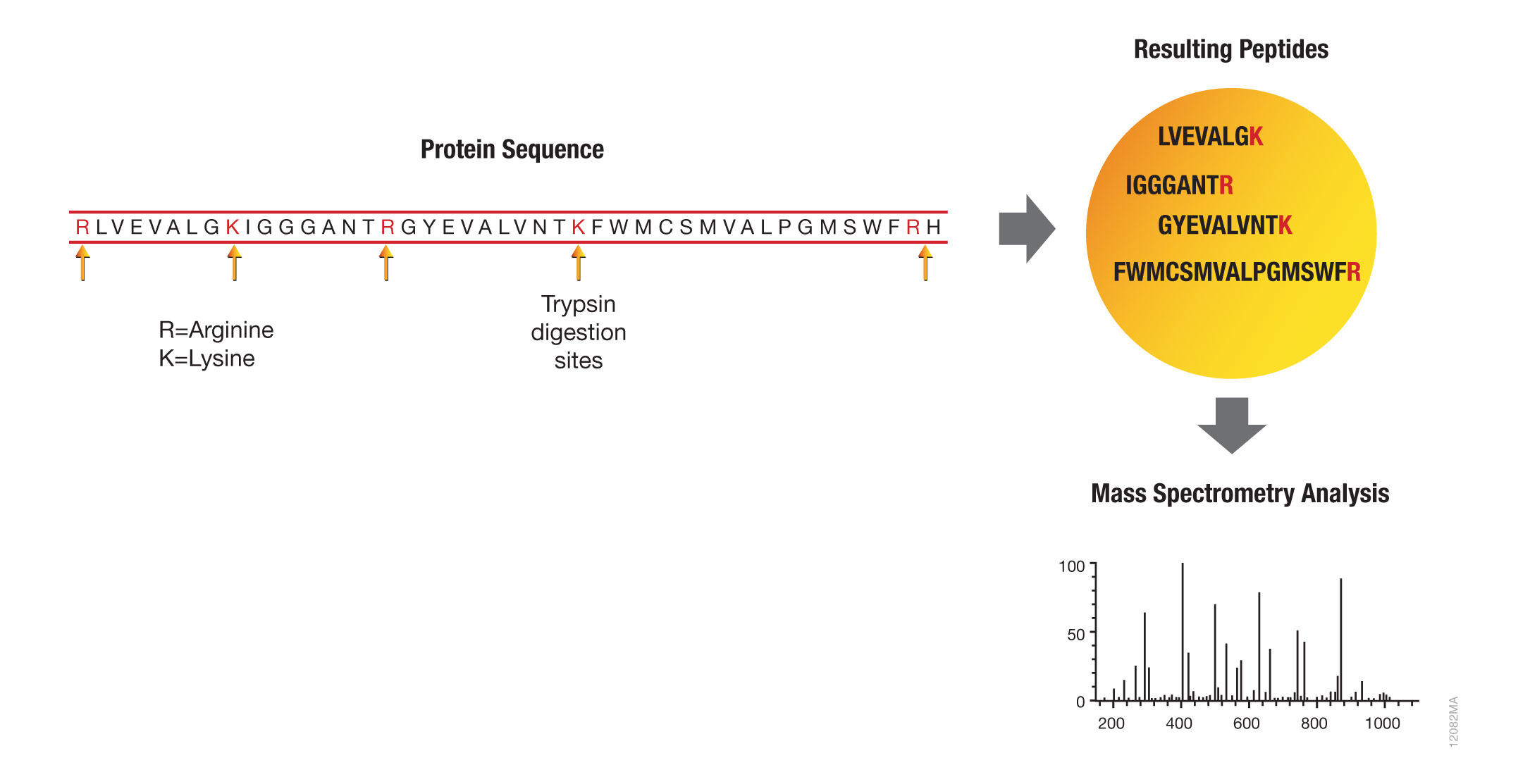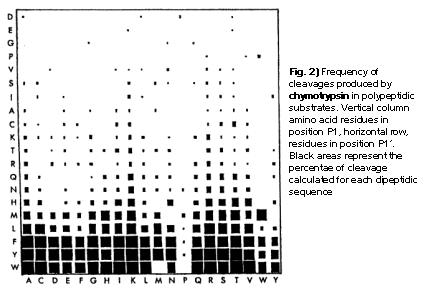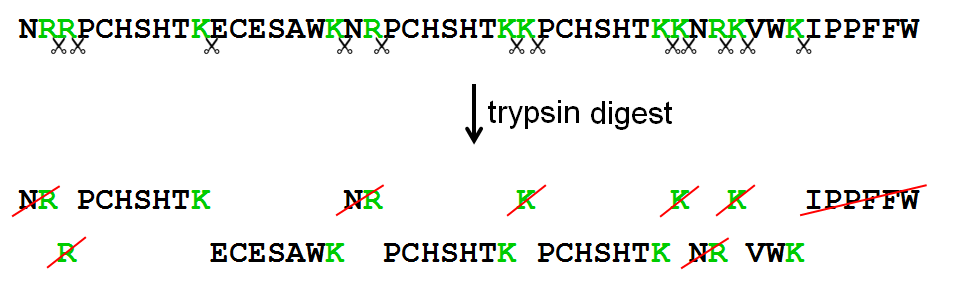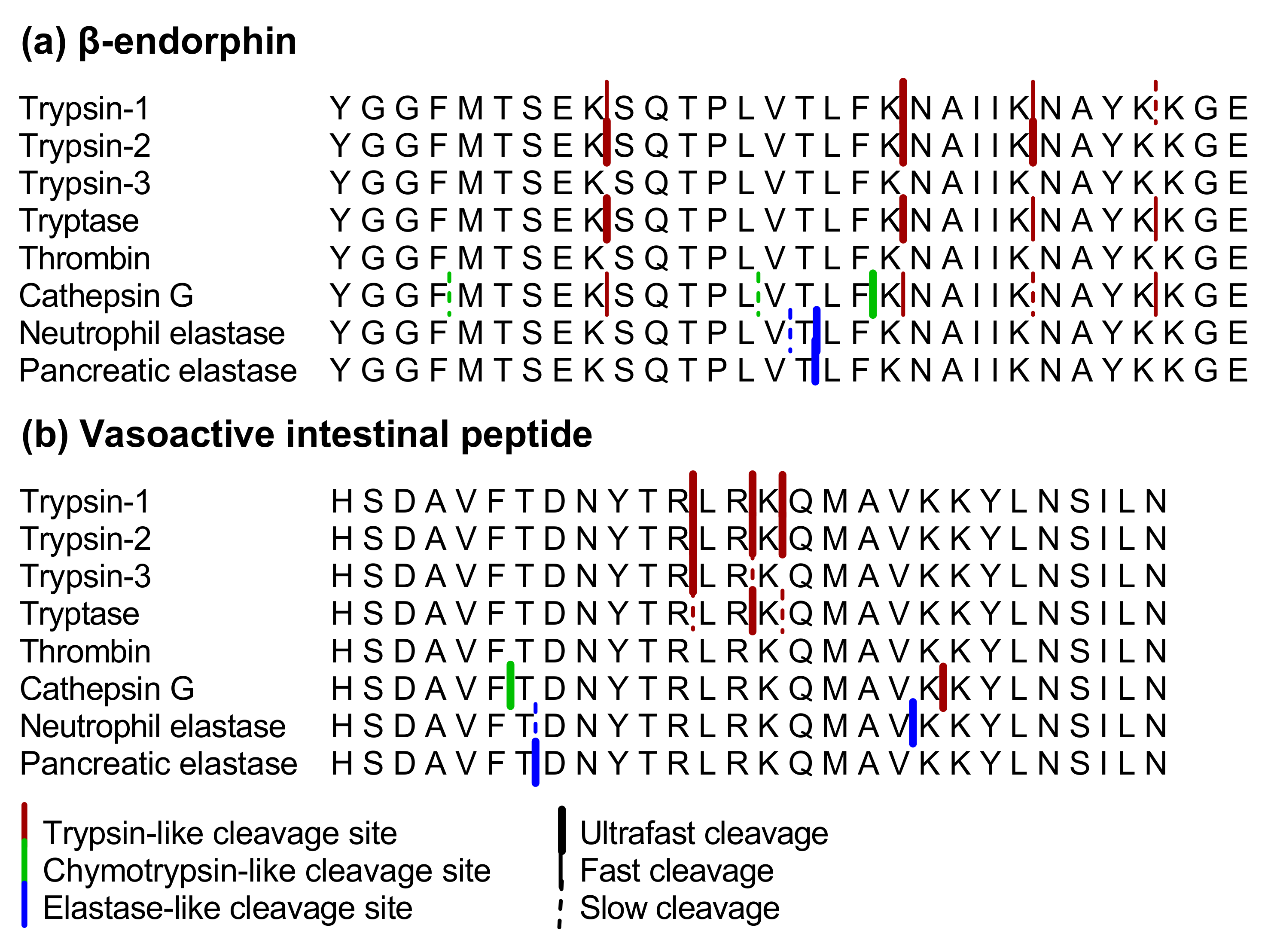
IJMS | Free Full-Text | Proteolytic Cleavage of Bioactive Peptides and Protease-Activated Receptors in Acute and Post-Colitis
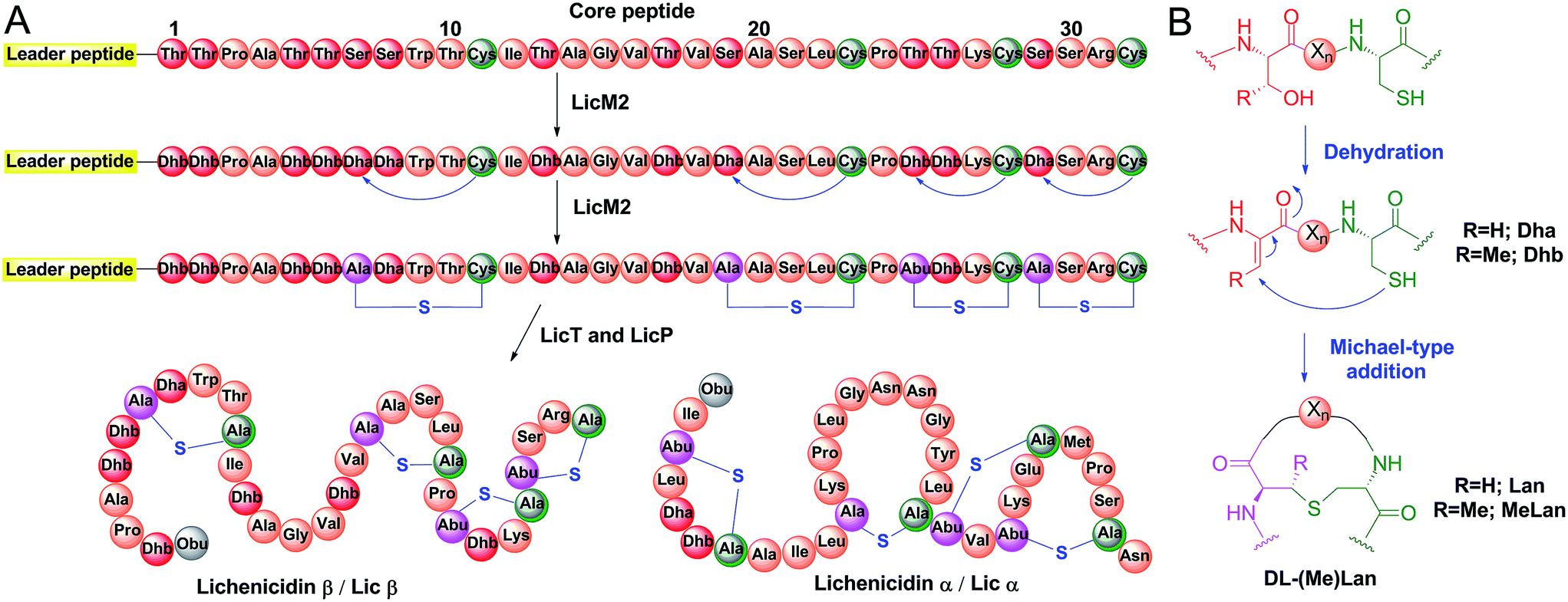
Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage - Chemical Science (RSC Publishing) DOI:10.1039/C5SC02329G
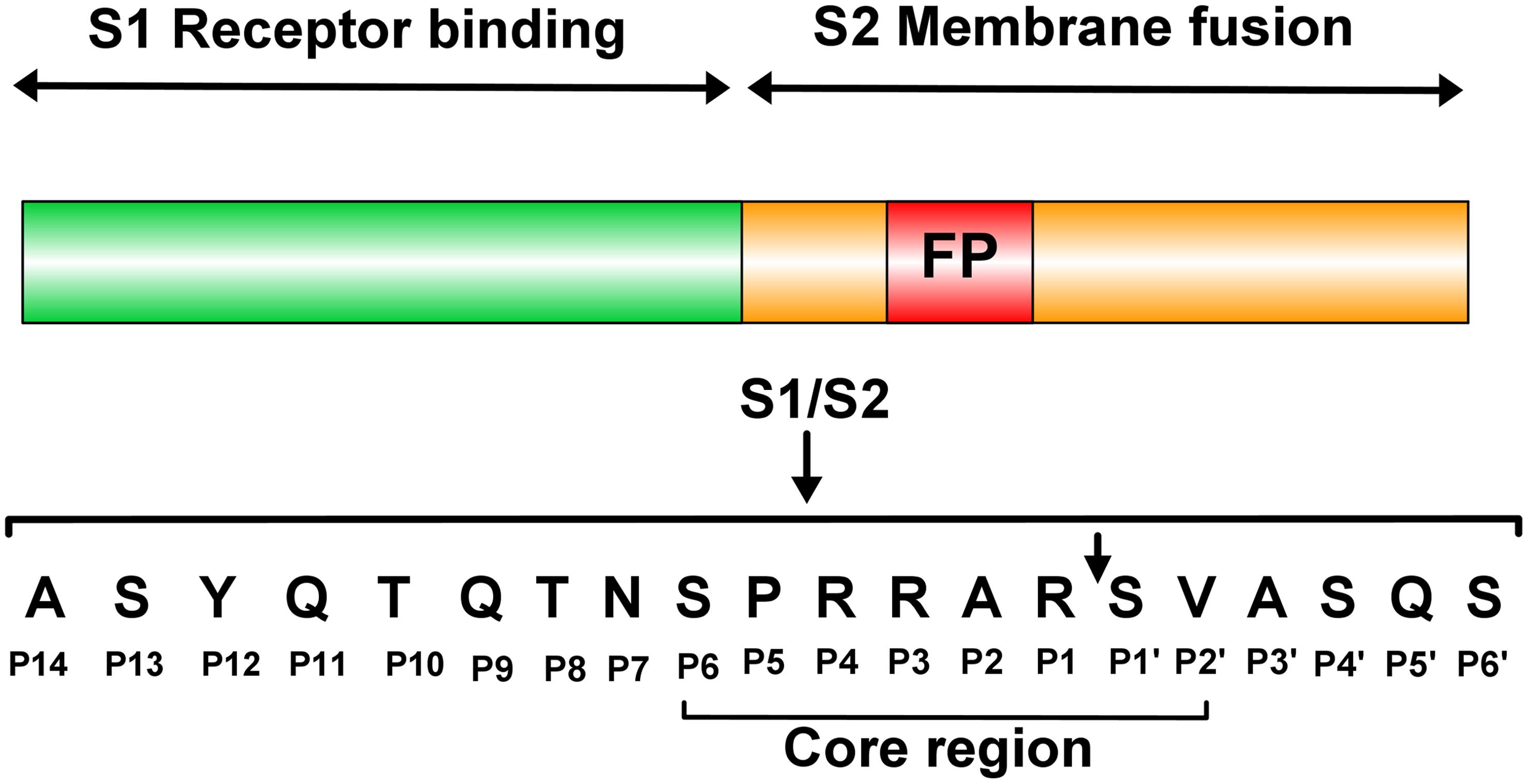
Hypertonic Saline and Aprotinin Inhibit Furin and Nasal Protease to Reduce SARS-CoV-2 Specific Furin Site Cleavage Activity

Amino acid sequence specifications for proteolytic cleavage by trypsin,... | Download Scientific Diagram

Protease cleavage site fingerprinting by label‐free in‐gel degradomics reveals pH‐dependent specificity switch of legumain | The EMBO Journal

Predominant cleavage of proteins N-terminal to serines and threonines using scandium(III) triflate | SpringerLink

Insight into Trypsin Miscleavage: Comparison of Kinetic Constants of Problematic Peptide Sequences | Analytical Chemistry

Proteomic Identification of protease Cleavage Sites (PICS) workflow.... | Download Scientific Diagram
![Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram](https://www.researchgate.net/publication/13465969/figure/fig1/AS:602031375003649@1520546873782/Sequence-of-HRPc-1-The-trypsin-cleavage-sites-are-indicated-by-bold-letters-The.png)
Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram

Schematic representation of the major trypsin cleavage sites and one... | Download Scientific Diagram