Global Sequencing of Proteolytic Cleavage Sites in Apoptosis by Specific Labeling of Protein N Termini: Cell

Nipah virus fusion protein: Influence of cleavage site mutations on the cleavability by cathepsin L, trypsin and furin | Semantic Scholar

PAR2 N-terminus with major cleavage sites identified. N-terminus of... | Download Scientific Diagram
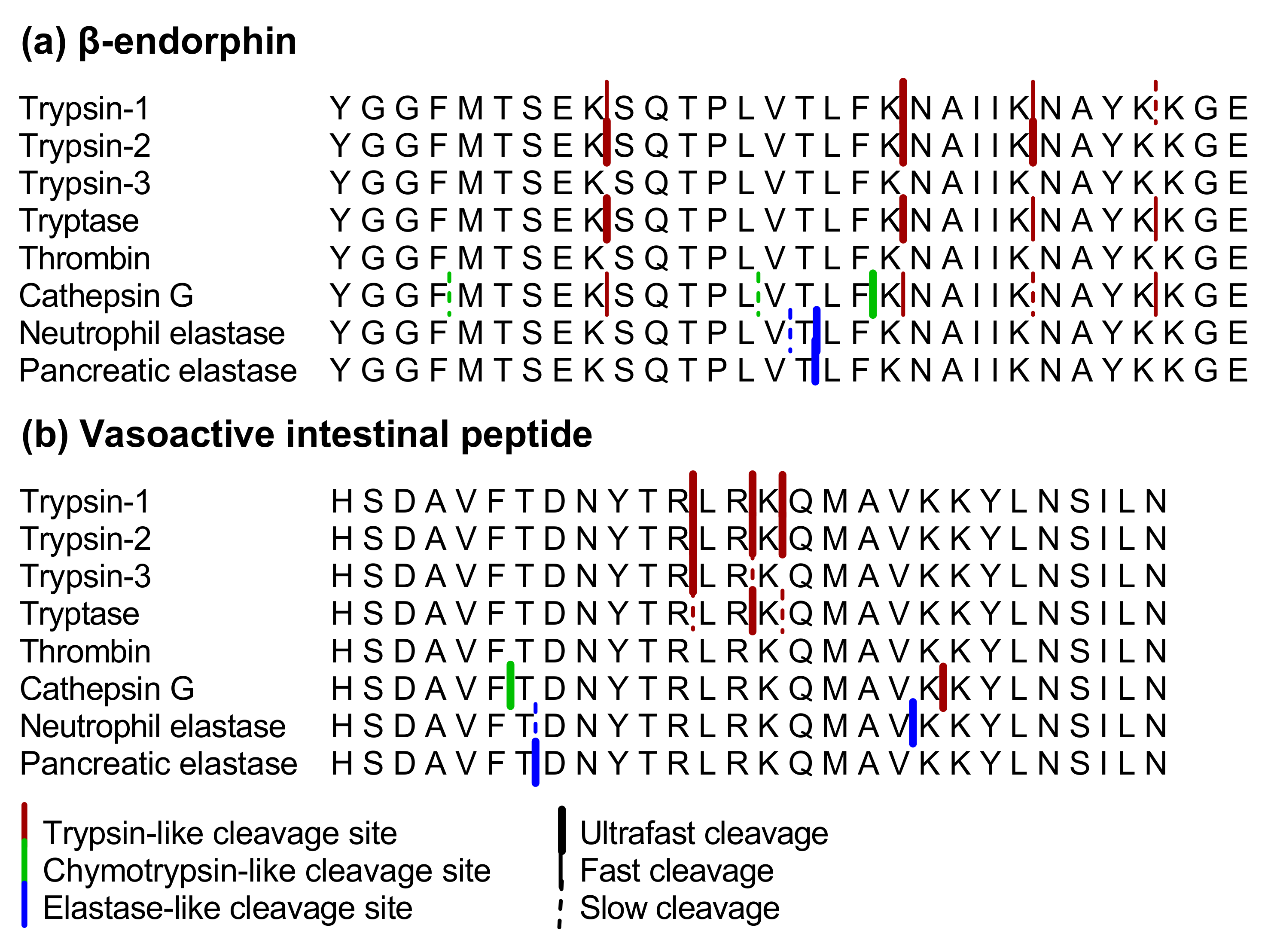
IJMS | Free Full-Text | Proteolytic Cleavage of Bioactive Peptides and Protease-Activated Receptors in Acute and Post-Colitis

Substrate cleavage specificity of legumain, GluC, and trypsin. IceLogos... | Download Scientific Diagram
![Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram](https://www.researchgate.net/publication/13465969/figure/fig1/AS:602031375003649@1520546873782/Sequence-of-HRPc-1-The-trypsin-cleavage-sites-are-indicated-by-bold-letters-The.png)
Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram
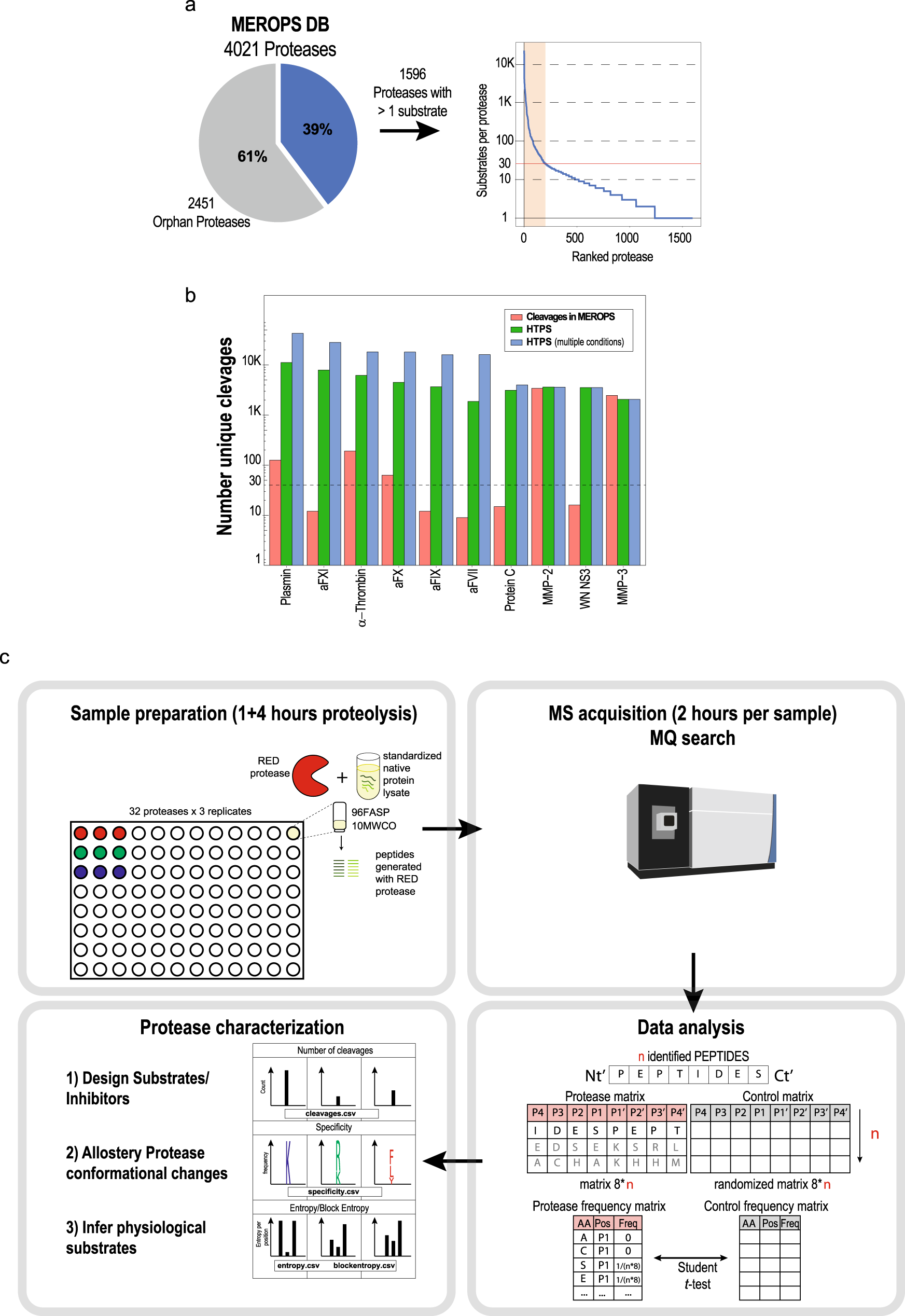
Mapping specificity, cleavage entropy, allosteric changes and substrates of blood proteases in a high-throughput screen | Nature Communications
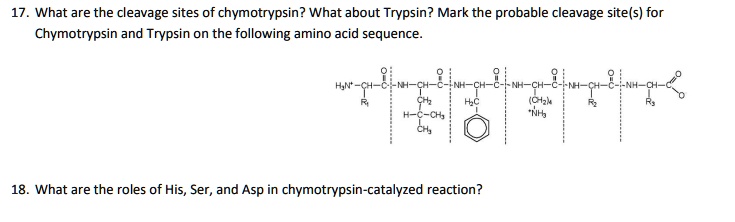
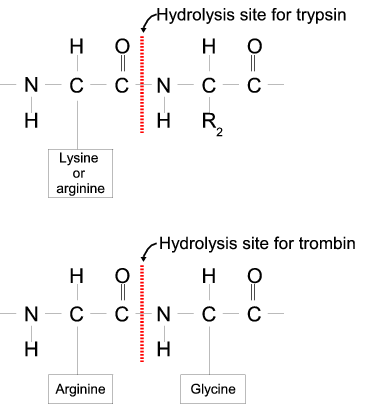
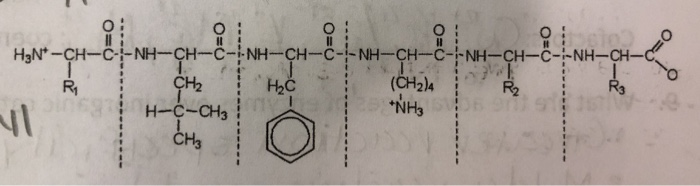
SOLVED: What are the cleavage sites of chymotrypsin? What about Trypsin? Mark the probable cleavage site(s) for Chymotrypsin and Trypsin on the following amino acid sequence: NH-CH (Czk "NI6 What are the

Protease cleavage site fingerprinting by label‐free in‐gel degradomics reveals pH‐dependent specificity switch of legumain | The EMBO Journal

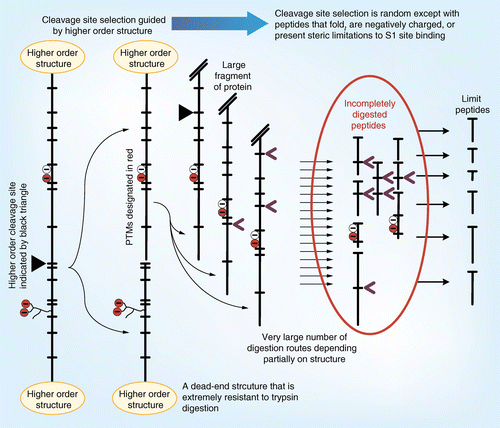
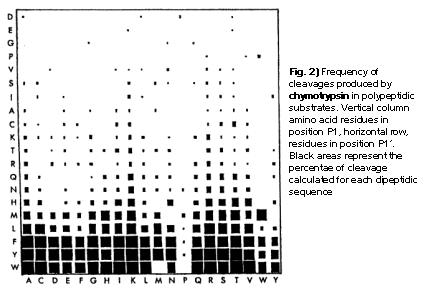
Marked difference in efficiency of the digestive enzymes pepsin, trypsin, chymotrypsin, and pancreatic elastase to cleave tightly folded proteins

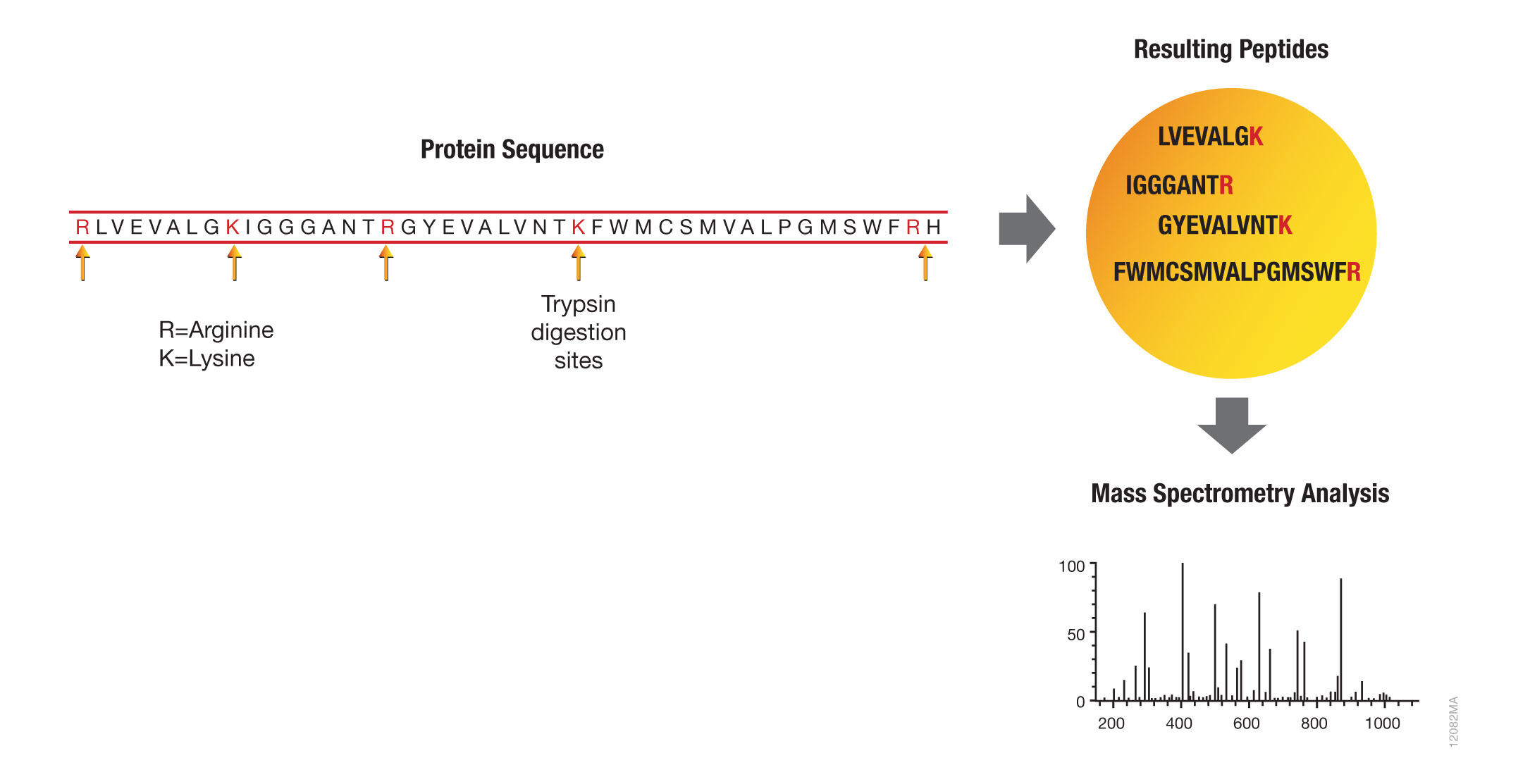
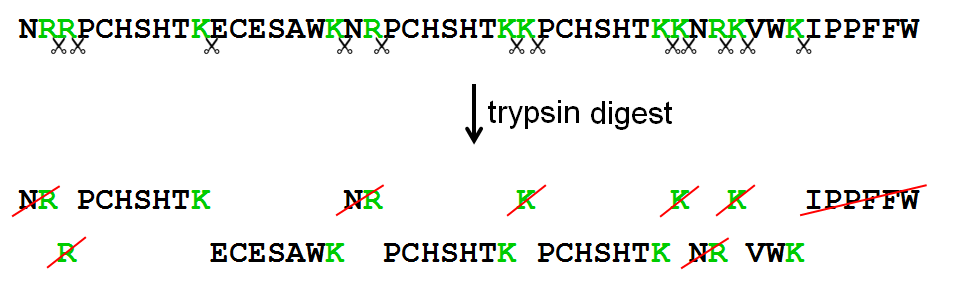
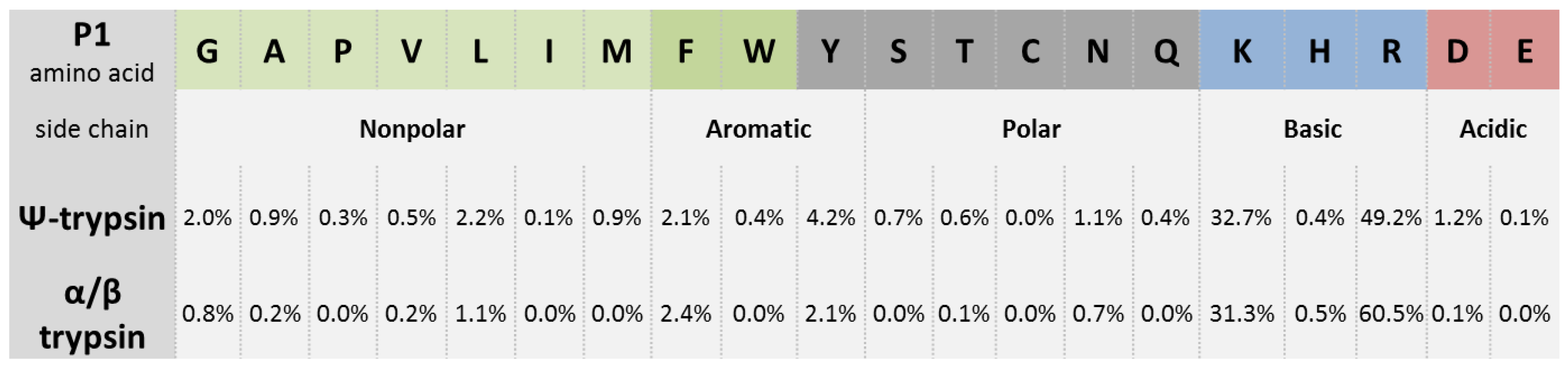
Cleaved and Missed Sites for Trypsin, Lys-C, and Lys-N Can Be Predicted with High Confidence on the Basis of Sequence Context | Journal of Proteome Research