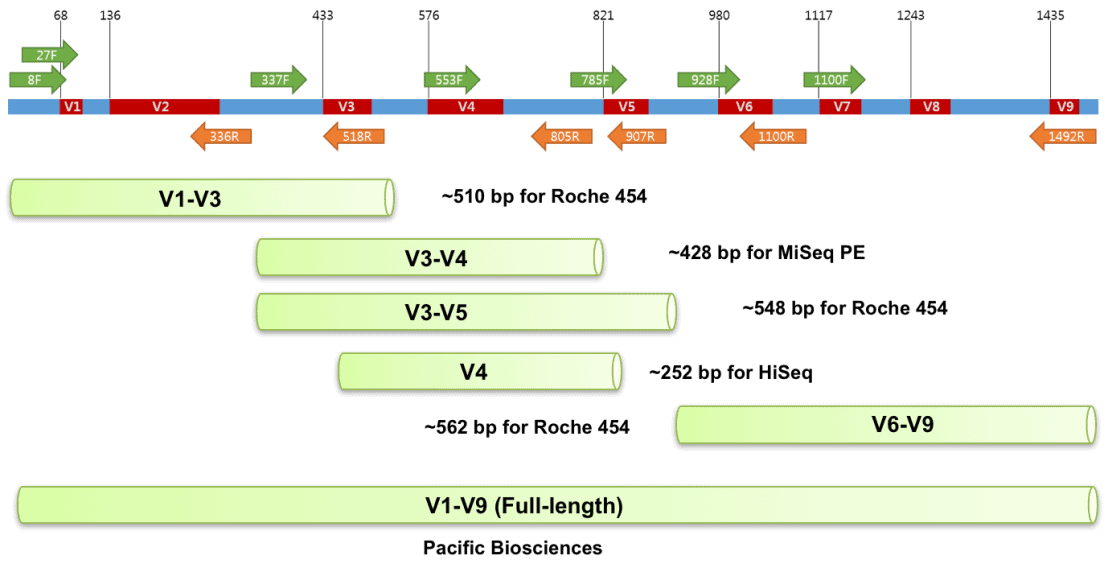
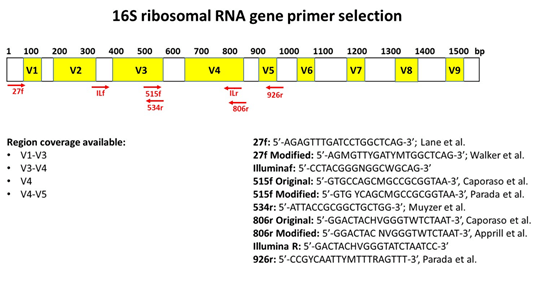
Positions of primer sequences (arrows) and 16S rRNA gene regions (bars)... | Download Scientific Diagram
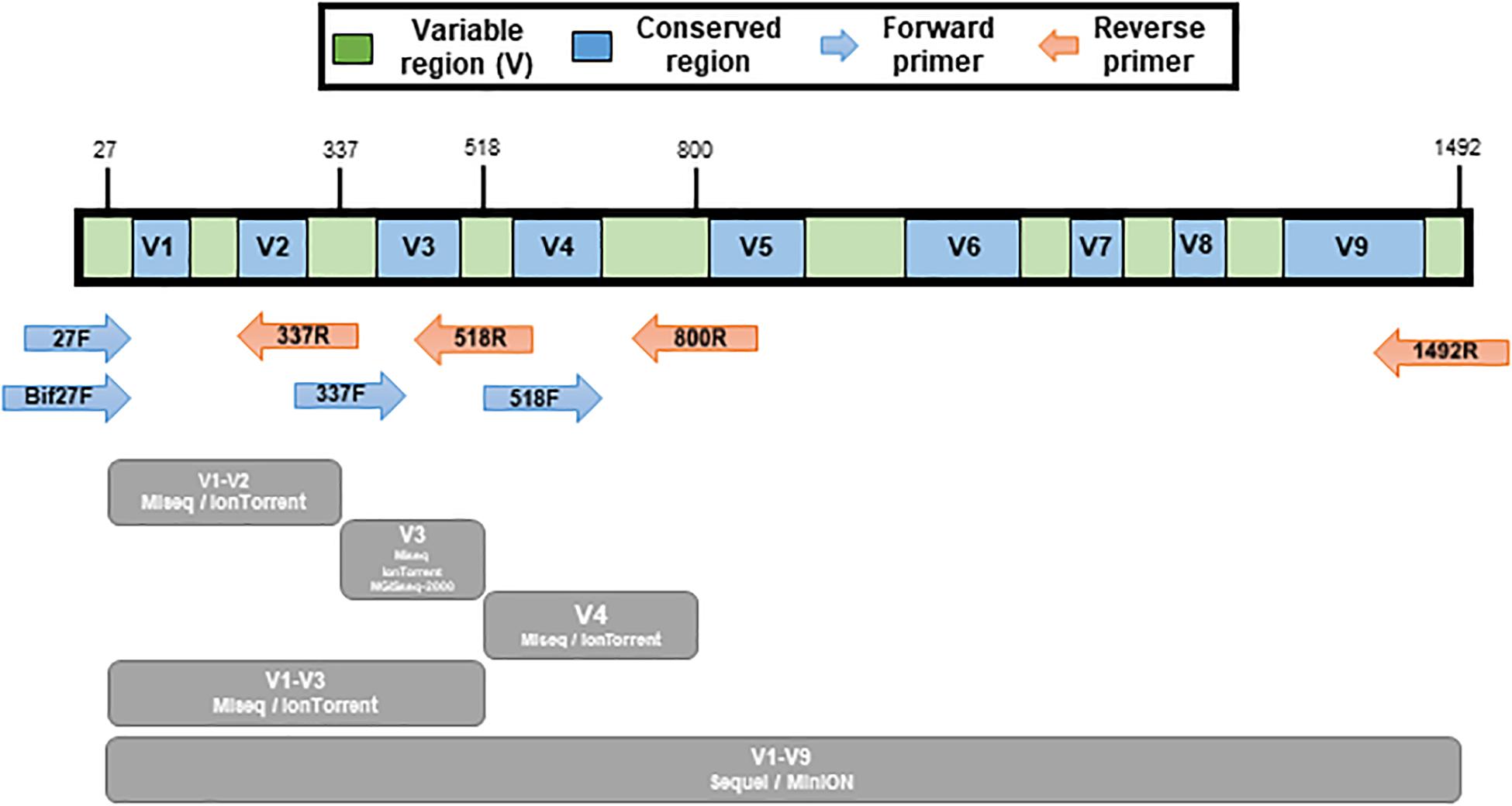
Frontiers | Comparison of 16S rRNA Gene Based Microbial Profiling Using Five Next-Generation Sequencers and Various Primers
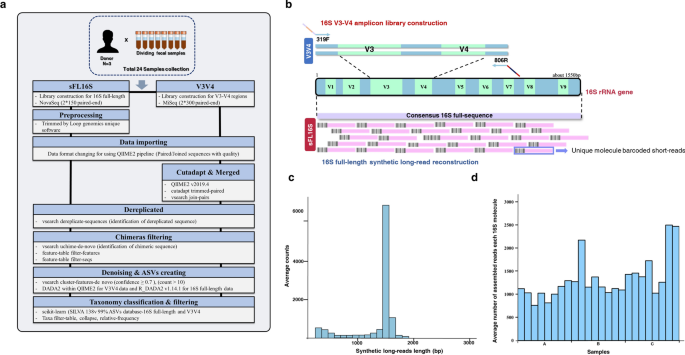
The effect of taxonomic classification by full-length 16S rRNA sequencing with a synthetic long-read technology | Scientific Reports
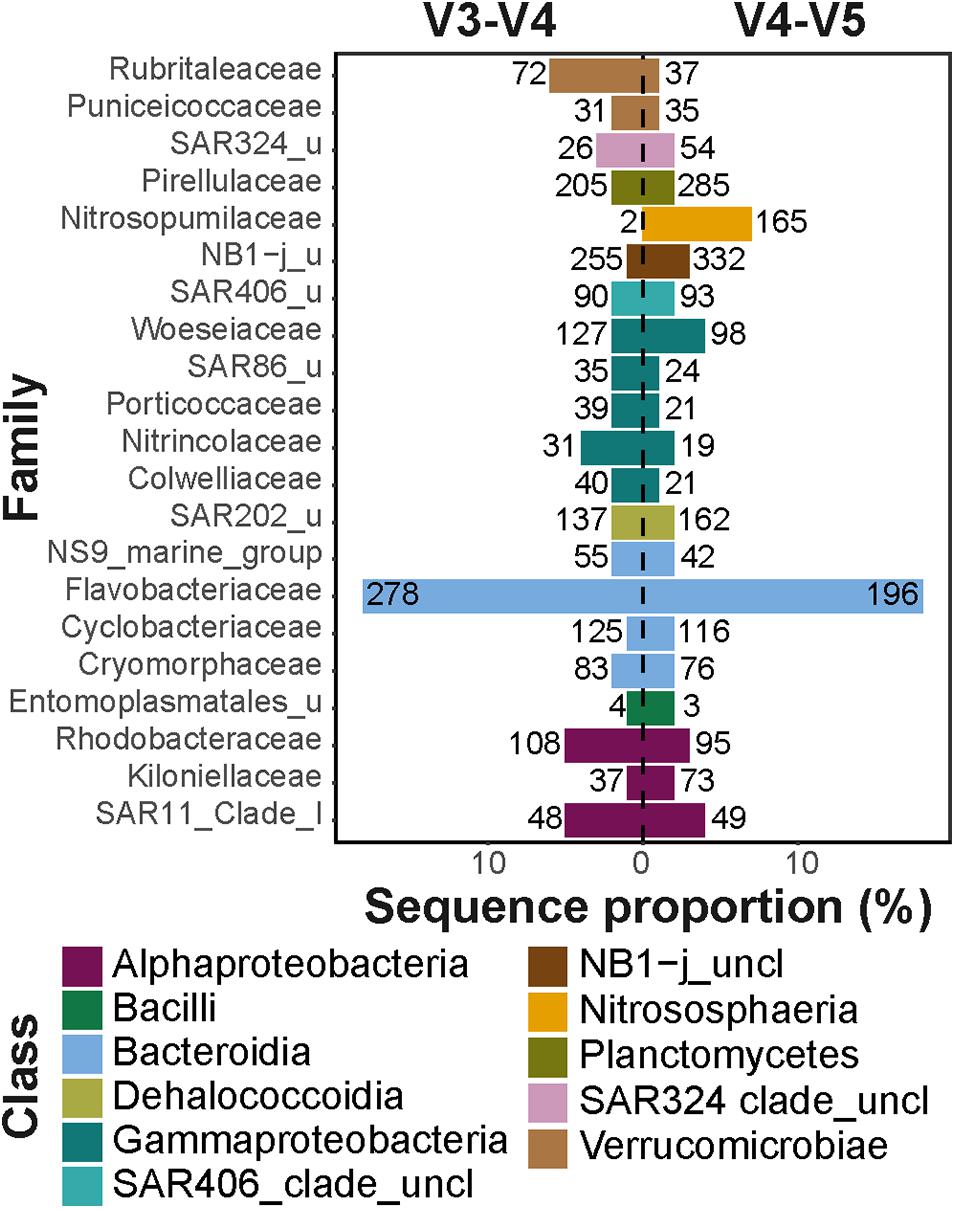
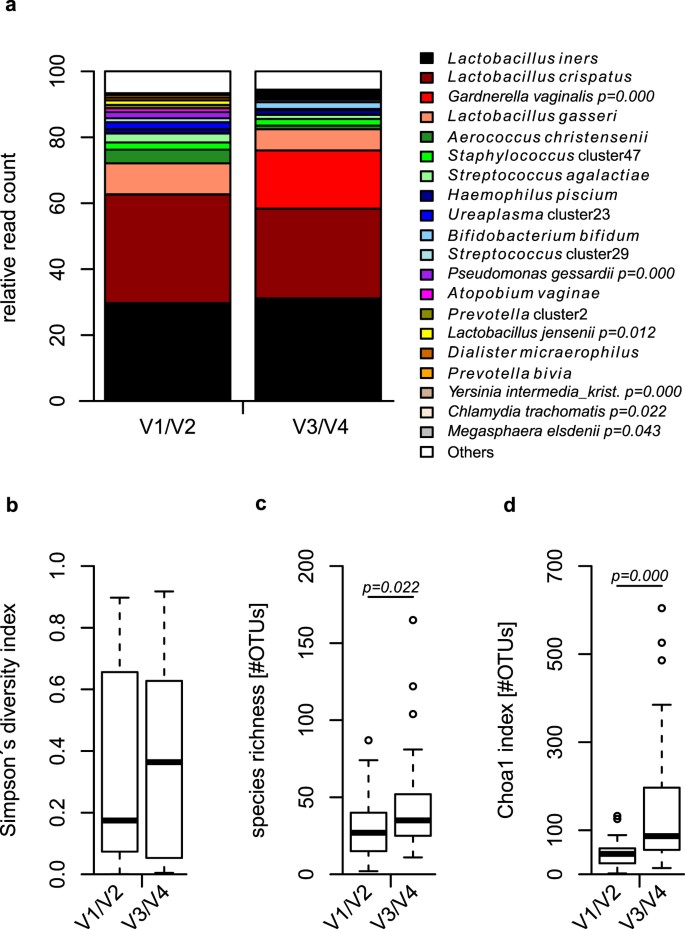
Frontiers | Comparison of Two 16S rRNA Primers (V3–V4 and V4–V5) for Studies of Arctic Microbial Communities
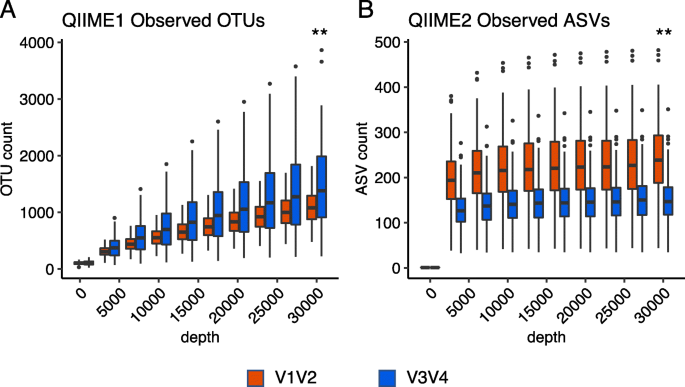
Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1–V2 and V3–V4 primer sets | BMC Genomics | Full Text

Schematic diagram of 16S rDNA sequencing workflow for both Run 1 and... | Download Scientific Diagram

List of modified Illumina V3-V4 primers and resulting read counts from... | Download Scientific Diagram
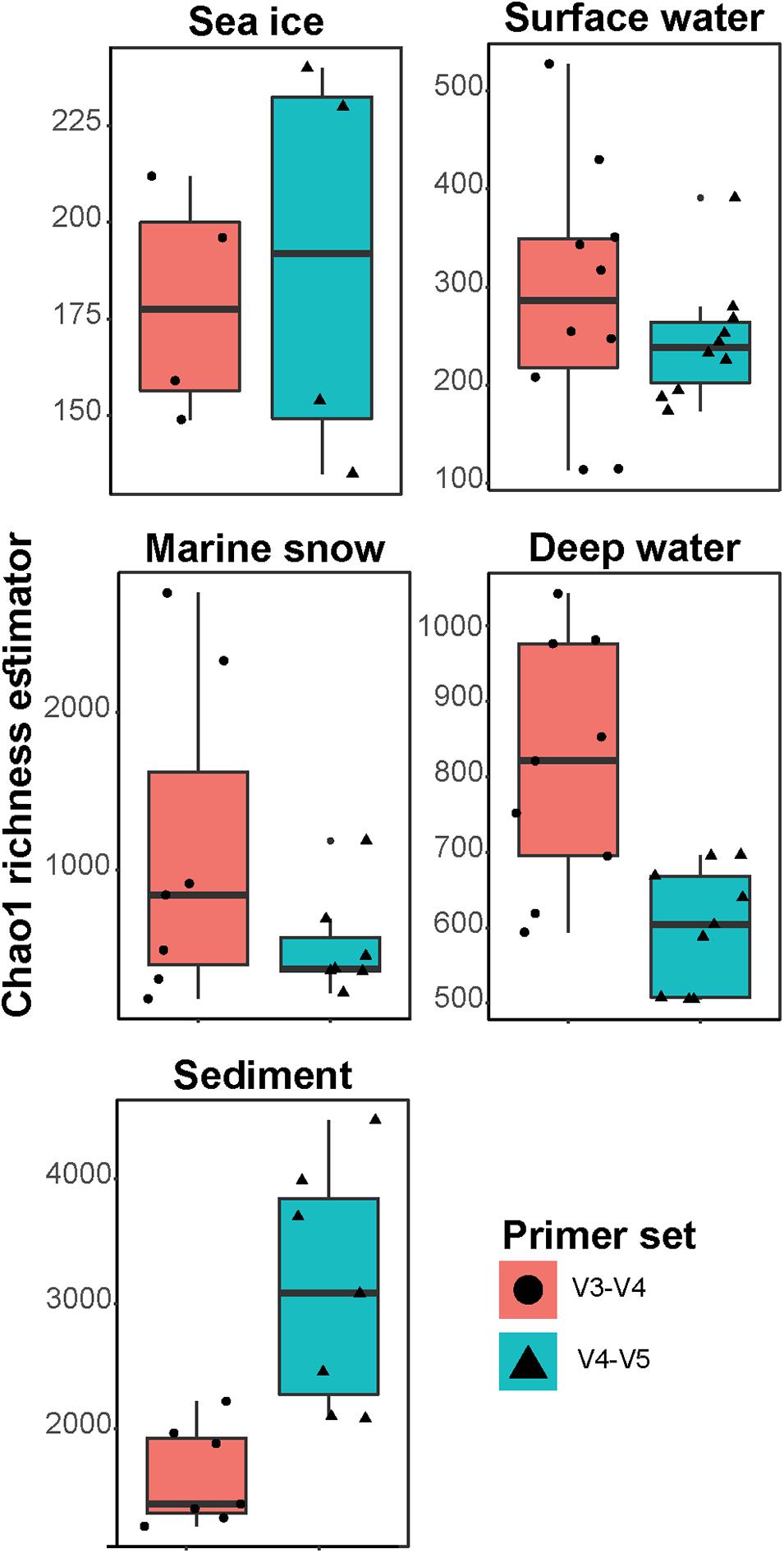
Frontiers | Comparison of Two 16S rRNA Primers (V3–V4 and V4–V5) for Studies of Arctic Microbial Communities

Reproducibility and repeatability of six high-throughput 16S rDNA sequencing protocols for microbiota profiling - ScienceDirect

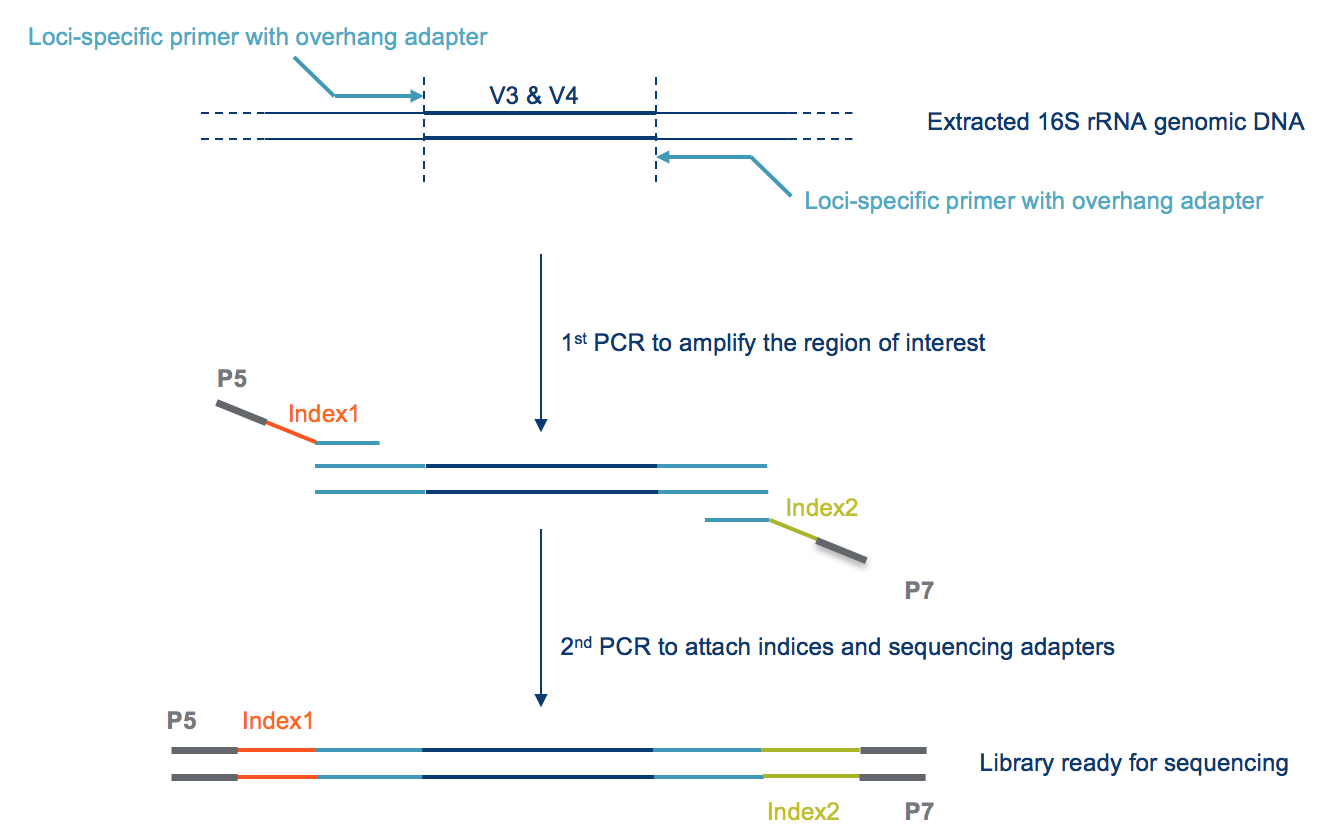
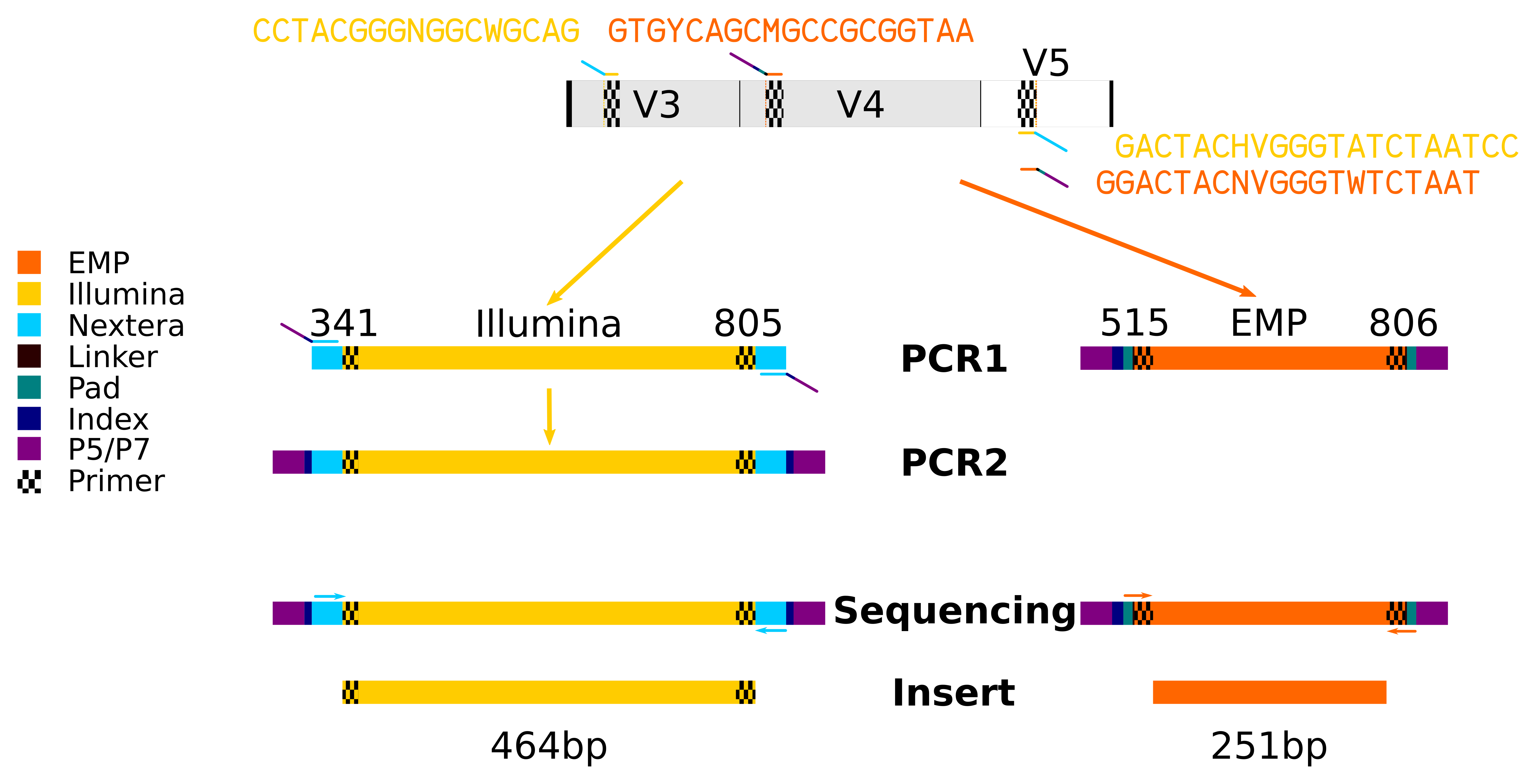
Evaluation of Compatibility of 16S rRNA V3V4 and V4 Amplicon Libraries for Clinical Microbiome Profiling | bioRxiv
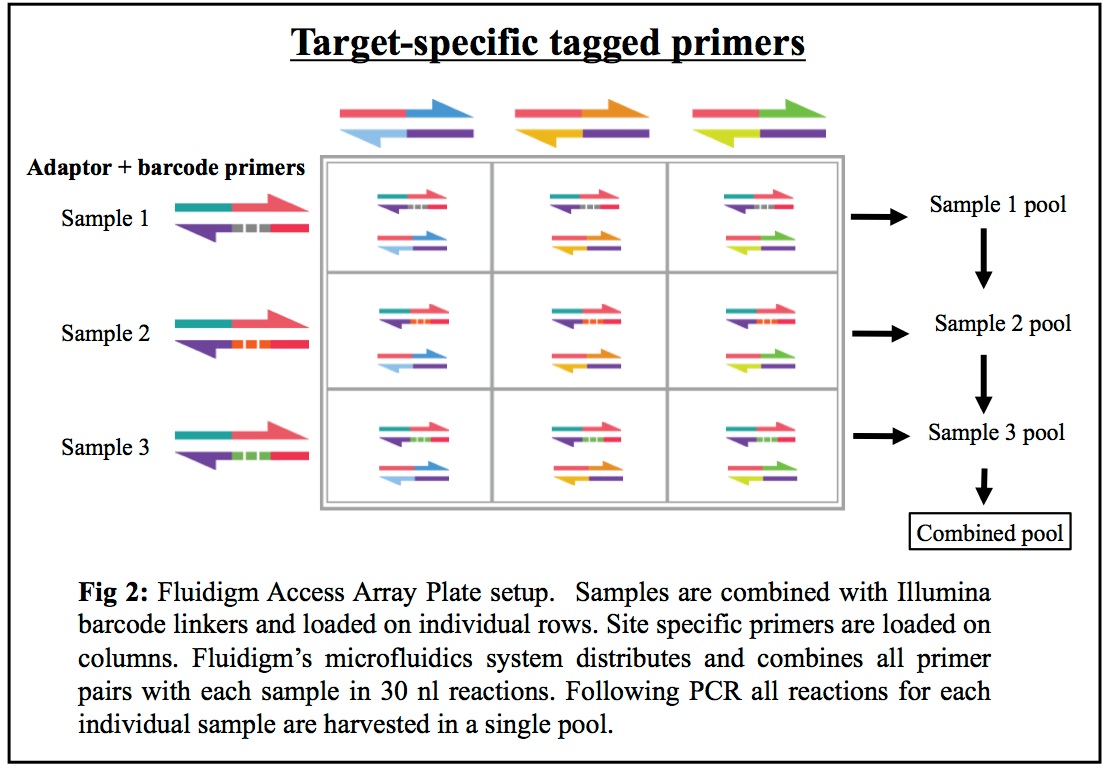
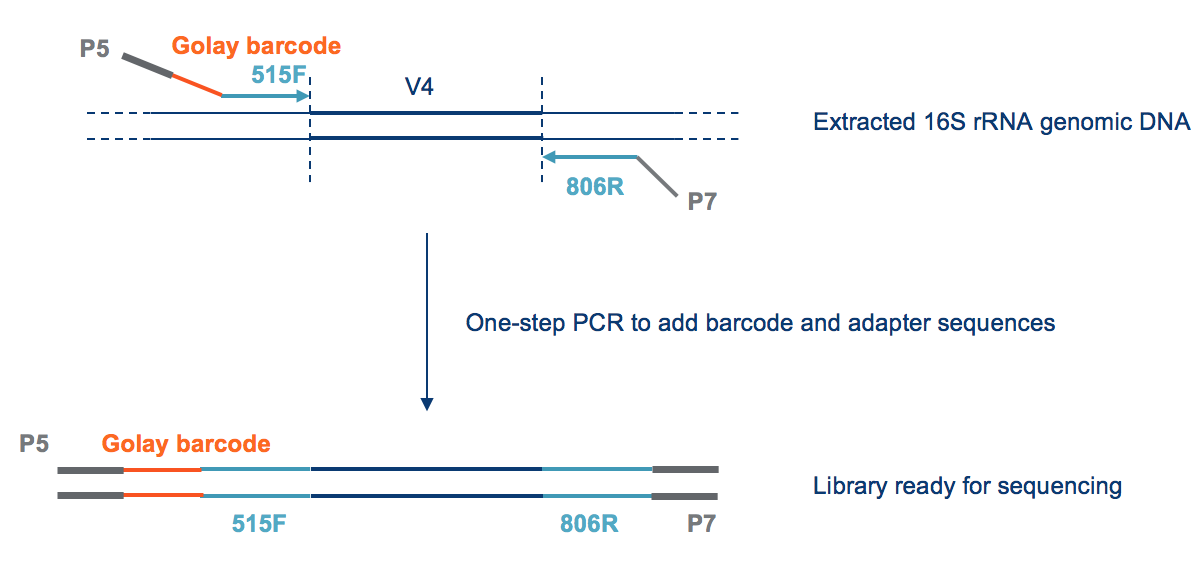
HTDNA: Services/Equipment: Preparation of 16S rRNA (and all other) Amplicons for Illumina Sequencing | Biotech

Evaluation of Compatibility of 16S rRNA V3V4 and V4 Amplicon Libraries for Clinical Microbiome Profiling | bioRxiv

Better skin microbiome analyses using new 16S V4 region primers developed by Microbiome Insights' scientific team — Microbiome Insights